Blockade of pachytene piRNA biogenesis reveals a novel requirement for maintaining post-meiotic germline genome integrity
- PMID: 23166510
- PMCID: PMC3499362
- DOI: 10.1371/journal.pgen.1003038
Blockade of pachytene piRNA biogenesis reveals a novel requirement for maintaining post-meiotic germline genome integrity
Abstract
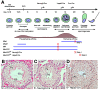
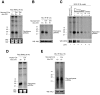
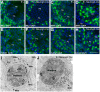
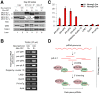
Piwi-interacting RNAs are a diverse class of small non-coding RNAs implicated in the silencing of transposable elements and the safeguarding of genome integrity. In mammals, male germ cells express two genetically and developmentally distinct populations of piRNAs at the pre-pachytene and pachytene stages of meiosis, respectively. Pre-pachytene piRNAs are mostly derived from retrotransposons and required for their silencing. In contrast, pachytene piRNAs originate from ~3,000 genomic clusters, and their biogenesis and function remain enigmatic. Here, we report that conditional inactivation of the putative RNA helicase MOV10L1 in mouse spermatocytes produces a specific loss of pachytene piRNAs, significant accumulation of pachytene piRNA precursor transcripts, and unusual polar conglomeration of Piwi proteins with mitochondria. Pachytene piRNA-deficient spermatocytes progress through meiosis without derepression of LINE1 retrotransposons, but become arrested at the post-meiotic round spermatid stage with massive DNA damage. Our results demonstrate that MOV10L1 acts upstream of Piwi proteins in the primary processing of pachytene piRNAs and suggest that, distinct from pre-pachytene piRNAs, pachytene piRNAs fulfill a unique function in maintaining post-meiotic genome integrity.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
yama, a mutant allele of Mov10l1, disrupts retrotransposon silencing and piRNA biogenesis.PLoS Genet. 2021 Feb 26;17(2):e1009265. doi: 10.1371/journal.pgen.1009265. eCollection 2021 Feb. PLoS Genet. 2021. PMID: 33635934 Free PMC article.
-
Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway.Proc Natl Acad Sci U S A. 2010 Jun 29;107(26):11841-6. doi: 10.1073/pnas.1003953107. Epub 2010 Jun 1. Proc Natl Acad Sci U S A. 2010. PMID: 20534472 Free PMC article.
-
MOV10L1 is necessary for protection of spermatocytes against retrotransposons by Piwi-interacting RNAs.Proc Natl Acad Sci U S A. 2010 Jun 29;107(26):11847-52. doi: 10.1073/pnas.1007158107. Epub 2010 Jun 14. Proc Natl Acad Sci U S A. 2010. PMID: 20547853 Free PMC article.
-
MOV10L1 in piRNA processing and gene silencing of retrotransposons during spermatogenesis.Reproduction. 2015 May;149(5):R229-35. doi: 10.1530/REP-14-0569. Epub 2015 Feb 9. Reproduction. 2015. PMID: 25667429 Review.
-
Enigmatic Pachytene PIWI-Interacting RNAs.Genome Biol Evol. 2024 Oct 9;16(10):evae162. doi: 10.1093/gbe/evae162. Genome Biol Evol. 2024. PMID: 39056586 Free PMC article. Review.
Cited by
-
Mitochondria Associated Germinal Structures in Spermatogenesis: piRNA Pathway Regulation and Beyond.Cells. 2020 Feb 10;9(2):399. doi: 10.3390/cells9020399. Cells. 2020. PMID: 32050598 Free PMC article. Review.
-
Roles of epigenome in mammalian spermatogenesis.Reprod Med Biol. 2013 Aug 22;13(2):59-69. doi: 10.1007/s12522-013-0167-8. eCollection 2014 Apr. Reprod Med Biol. 2013. PMID: 29699150 Free PMC article. Review.
-
yama, a mutant allele of Mov10l1, disrupts retrotransposon silencing and piRNA biogenesis.PLoS Genet. 2021 Feb 26;17(2):e1009265. doi: 10.1371/journal.pgen.1009265. eCollection 2021 Feb. PLoS Genet. 2021. PMID: 33635934 Free PMC article.
-
Mitochondrial membrane-based initial separation of MIWI and MILI functions during pachytene piRNA biogenesis.Nucleic Acids Res. 2019 Mar 18;47(5):2594-2608. doi: 10.1093/nar/gky1281. Nucleic Acids Res. 2019. PMID: 30590800 Free PMC article.
-
Single-cell RNA-seq uncovers dynamic processes orchestrated by RNA-binding protein DDX43 in chromatin remodeling during spermiogenesis.Nat Commun. 2023 Apr 29;14(1):2499. doi: 10.1038/s41467-023-38199-w. Nat Commun. 2023. PMID: 37120627 Free PMC article.
References
-
- Siomi MC, Sato K, Pezic D, Aravin AA (2011) PIWI-interacting small RNAs: The vanguard of genome defence. Nat Rev Mol Cell Biol 12: 246–258. - PubMed
-
- Pillai RS, Chuma S (2012) piRNAs and their involvement in male germline development in mice. Dev Growth Differ - PubMed
-
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, et al. (2001) Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol 11: 1017–1027. - PubMed
-
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, et al. (2007) Discrete small RNA-generating loci as master regulators of transposon activity in drosophila. Cell 128: 1089–1103. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

