Genome, functional gene annotation, and nuclear transformation of the heterokont oleaginous alga Nannochloropsis oceanica CCMP1779
- PMID: 23166516
- PMCID: PMC3499364
- DOI: 10.1371/journal.pgen.1003064
Genome, functional gene annotation, and nuclear transformation of the heterokont oleaginous alga Nannochloropsis oceanica CCMP1779
Erratum in
-
Correction: Genome, Functional Gene Annotation, and Nuclear Transformation of the Heterokont Oleaginous Alga Nannochloropsis oceanica CCMP1779.PLoS Genet. 2017 May 23;13(5):e1006802. doi: 10.1371/journal.pgen.1006802. eCollection 2017 May. PLoS Genet. 2017. PMID: 28542203 Free PMC article.
Abstract
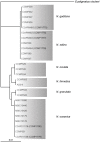
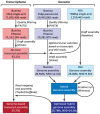
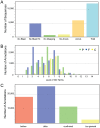
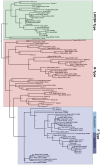
Unicellular marine algae have promise for providing sustainable and scalable biofuel feedstocks, although no single species has emerged as a preferred organism. Moreover, adequate molecular and genetic resources prerequisite for the rational engineering of marine algal feedstocks are lacking for most candidate species. Heterokonts of the genus Nannochloropsis naturally have high cellular oil content and are already in use for industrial production of high-value lipid products. First success in applying reverse genetics by targeted gene replacement makes Nannochloropsis oceanica an attractive model to investigate the cell and molecular biology and biochemistry of this fascinating organism group. Here we present the assembly of the 28.7 Mb genome of N. oceanica CCMP1779. RNA sequencing data from nitrogen-replete and nitrogen-depleted growth conditions support a total of 11,973 genes, of which in addition to automatic annotation some were manually inspected to predict the biochemical repertoire for this organism. Among others, more than 100 genes putatively related to lipid metabolism, 114 predicted transcription factors, and 109 transcriptional regulators were annotated. Comparison of the N. oceanica CCMP1779 gene repertoire with the recently published N. gaditana genome identified 2,649 genes likely specific to N. oceanica CCMP1779. Many of these N. oceanica-specific genes have putative orthologs in other species or are supported by transcriptional evidence. However, because similarity-based annotations are limited, functions of most of these species-specific genes remain unknown. Aside from the genome sequence and its analysis, protocols for the transformation of N. oceanica CCMP1779 are provided. The availability of genomic and transcriptomic data for Nannochloropsis oceanica CCMP1779, along with efficient transformation protocols, provides a blueprint for future detailed gene functional analysis and genetic engineering of Nannochloropsis species by a growing academic community focused on this genus.
Conflict of interest statement
CB and KKN are on the advisory board of Aurora Alga. AV was supported in part by a grant from Aurora Alga.
Figures
Similar articles
-
Transcriptional coordination of physiological responses in Nannochloropsis oceanica CCMP1779 under light/dark cycles.Plant J. 2015 Sep;83(6):1097-113. doi: 10.1111/tpj.12944. Epub 2015 Aug 20. Plant J. 2015. PMID: 26216534
-
Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropis gaditana.Nat Commun. 2012 Feb 21;3:686. doi: 10.1038/ncomms1688. Nat Commun. 2012. PMID: 22353717 Free PMC article.
-
Correction: Genome, Functional Gene Annotation, and Nuclear Transformation of the Heterokont Oleaginous Alga Nannochloropsis oceanica CCMP1779.PLoS Genet. 2017 May 23;13(5):e1006802. doi: 10.1371/journal.pgen.1006802. eCollection 2017 May. PLoS Genet. 2017. PMID: 28542203 Free PMC article.
-
Advanced genetic tools enable synthetic biology in the oleaginous microalgae Nannochloropsis sp.Plant Cell Rep. 2018 Oct;37(10):1383-1399. doi: 10.1007/s00299-018-2270-0. Epub 2018 Mar 6. Plant Cell Rep. 2018. PMID: 29511798 Review.
-
Biochemistry and Biotechnology of Lipid Accumulation in the Microalga Nannochloropsis oceanica.J Agric Food Chem. 2022 Sep 21;70(37):11500-11509. doi: 10.1021/acs.jafc.2c05309. Epub 2022 Sep 9. J Agric Food Chem. 2022. PMID: 36083864 Review.
Cited by
-
Characterization of Hydrogen Metabolism in the Multicellular Green Alga Volvox carteri.PLoS One. 2015 Apr 30;10(4):e0125324. doi: 10.1371/journal.pone.0125324. eCollection 2015. PLoS One. 2015. PMID: 25927230 Free PMC article.
-
The role of pyruvate hub enzymes in supplying carbon precursors for fatty acid synthesis in photosynthetic microalgae.Photosynth Res. 2015 Sep;125(3):407-22. doi: 10.1007/s11120-015-0136-7. Epub 2015 Apr 7. Photosynth Res. 2015. PMID: 25846135
-
Integration of proteome and transcriptome refines key molecular processes underlying oil production in Nannochloropsis oceanica.Biotechnol Biofuels. 2020 Jun 18;13:109. doi: 10.1186/s13068-020-01748-2. eCollection 2020. Biotechnol Biofuels. 2020. PMID: 32565907 Free PMC article.
-
A Modeled High-Density Fed-Batch Culture Improves Biomass Growth and β-Glucans Accumulation in Microchloropsis salina.Plants (Basel). 2022 Nov 25;11(23):3229. doi: 10.3390/plants11233229. Plants (Basel). 2022. PMID: 36501269 Free PMC article.
-
Phosphorus-Induced Lipid Class Alteration Revealed by Lipidomic and Transcriptomic Profiling in Oleaginous Microalga Nannochloropsis sp. PJ12.Mar Drugs. 2019 Sep 3;17(9):519. doi: 10.3390/md17090519. Mar Drugs. 2019. PMID: 31484443 Free PMC article.
References
-
- Dismukes GC, Carrieri D, Bennette N, Ananyev GM, Posewitz MC (2008) Aquatic phototrophs: efficient alternatives to land-based crops for biofuels. Curr Opin Biotechnol 19: 235–240. - PubMed
-
- Wijffels RH, Barbosa MJ (2010) An outlook on microalgal biofuels. Science 329: 796–799. - PubMed
-
- Weyer KM, Bush DR, Darzins A, Willson BD (2010) Theoretical maximum algal oil production. Bioenergy Res 3: 204–213.
-
- Bowler C, Allen AE, Badger JH, Grimwood J, Jabbari K, et al. (2008) The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456: 239–244. - PubMed
-
- Siaut M, Heijde M, Mangogna M, Montsant A, Coesel S, et al. (2007) Molecular toolbox for studying diatom biology in Phaeodactylum tricornutum. Gene 406: 23–35. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

