Comparative analysis of the recently discovered hAT transposon TcBuster in human cells
- PMID: 23166581
- PMCID: PMC3499496
- DOI: 10.1371/journal.pone.0042666
Comparative analysis of the recently discovered hAT transposon TcBuster in human cells
Abstract
Background: Transposons are useful tools for creating transgenic organisms, insertional mutagenesis, and genome engineering. TcBuster, a novel hAT-family transposon system derived from the red flour beetle Tribolium castaneum, was shown to be highly active in previous studies in insect embryoes.
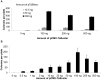
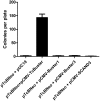
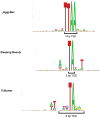
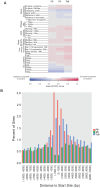
Methodology/principal findings: We tested TcBuster for its activity in human embryonic kidney 293 (HEK-293) cells. Excision footprints obtained from HEK-293 cells contained small insertions and deletions consistent with a hAT-type repair mechanism of hairpin formation and non-homologous end-joining. Genome-wide analysis of 23,417 piggyBac, 30,303 Sleeping Beauty, and 27,985 TcBuster integrations in HEK-293 cells revealed a uniquely different integration pattern when compared to other transposon systems with regards to genomic elements. TcBuster experimental conditions were optimized to assay TcBuster activity in HEK-293 cells by colony assay selection for a neomycin-containing transposon. Increasing transposon plasmid increased the number of colonies, whereas gene transfer activity dependent on codon-optimized transposase plasmid peaked at 100 ng with decreased colonies at the highest doses of transposase DNA. Expression of the related human proteins Buster1, Buster3, and SCAND3 in HEK-293 cells did not result in genomic integration of the TcBuster transposon. TcBuster, Tol2, and piggyBac were compared directly at different ratios of transposon to transposase and found to be approximately comparable while having their own ratio preferences.
Conclusions/significance: TcBuster was found to be highly active in mammalian HEK-293 cells and represents a promising tool for mammalian genome engineering.
Conflict of interest statement
Figures


Similar articles
-
A resurrected mammalian hAT transposable element and a closely related insect element are highly active in human cell culture.Proc Natl Acad Sci U S A. 2013 Feb 5;110(6):E478-87. doi: 10.1073/pnas.1121543109. Epub 2012 Oct 22. Proc Natl Acad Sci U S A. 2013. PMID: 23091042 Free PMC article.
-
Functional analysis of the catalytic triad of the hAT-family transposase TcBuster.Plasmid. 2021 Mar;114:102554. doi: 10.1016/j.plasmid.2021.102554. Epub 2021 Jan 18. Plasmid. 2021. PMID: 33476638 Free PMC article.
-
Genome-wide target profiling of piggyBac and Tol2 in HEK 293: pros and cons for gene discovery and gene therapy.BMC Biotechnol. 2011 Mar 30;11:28. doi: 10.1186/1472-6750-11-28. BMC Biotechnol. 2011. PMID: 21447194 Free PMC article.
-
Size matters: versatile use of PiggyBac transposons as a genetic manipulation tool.Mol Cell Biochem. 2011 Aug;354(1-2):301-9. doi: 10.1007/s11010-011-0832-3. Epub 2011 Apr 23. Mol Cell Biochem. 2011. PMID: 21516337 Review.
-
The Sleeping Beauty transposon toolbox.Methods Mol Biol. 2012;859:229-40. doi: 10.1007/978-1-61779-603-6_13. Methods Mol Biol. 2012. PMID: 22367875 Review.
Cited by
-
Sequencing methods and datasets to improve functional interpretation of sleeping beauty mutagenesis screens.BMC Genomics. 2014 Dec 19;15(1):1150. doi: 10.1186/1471-2164-15-1150. BMC Genomics. 2014. PMID: 25526783 Free PMC article.
-
An adaptable system for improving transposon-based gene expression in vivo via transient transgene repression.FASEB J. 2013 Sep;27(9):3753-62. doi: 10.1096/fj.13-232090. Epub 2013 Jun 10. FASEB J. 2013. PMID: 23752206 Free PMC article.
-
Decaying and expanding Erk gradients process memory of skeletal size during zebrafish fin regeneration.bioRxiv [Preprint]. 2025 Jan 23:2025.01.23.634576. doi: 10.1101/2025.01.23.634576. bioRxiv. 2025. PMID: 39896678 Free PMC article. Preprint.
-
Evolution of the clinical-stage hyperactive TcBuster transposase as a platform for robust non-viral production of adoptive cellular therapies.Mol Ther. 2024 Jun 5;32(6):1817-1834. doi: 10.1016/j.ymthe.2024.04.024. Epub 2024 Apr 16. Mol Ther. 2024. PMID: 38627969 Free PMC article.
-
A simplified transposon mutagenesis method to perform phenotypic forward genetic screens in cultured cells.BMC Genomics. 2019 Jun 17;20(1):497. doi: 10.1186/s12864-019-5888-6. BMC Genomics. 2019. PMID: 31208320 Free PMC article.
References
-
- Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA (2005) Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature 436: 272–276. - PubMed
-
- Ivics Z, Hackett PB, Plasterk RH, Izsvak Z (1997) Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell 91: 501–510. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

