Evolutionary dynamics of the interferon-induced transmembrane gene family in vertebrates
- PMID: 23166625
- PMCID: PMC3499546
- DOI: 10.1371/journal.pone.0049265
Evolutionary dynamics of the interferon-induced transmembrane gene family in vertebrates
Abstract
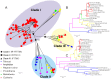
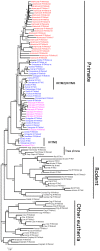
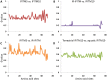
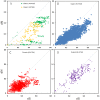
Vertebrate interferon-induced transmembrane (IFITM) genes have been demonstrated to have extensive and diverse functions, playing important roles in the evolution of vertebrates. Despite observance of their functionality, the evolutionary dynamics of this gene family are complex and currently unknown. Here, we performed detailed evolutionary analyses to unravel the evolutionary history of the vertebrate IFITM family. A total of 174 IFITM orthologous genes and 112 pseudogenes were identified from 27 vertebrate genome sequences. The vertebrate IFITM family can be divided into immunity-related IFITM (IR-IFITM), IFITM5 and IFITM10 sub-families in phylogeny, implying origins from three different progenitors. In general, vertebrate IFITM genes are located in two loci, one containing the IFITM10 gene, and the other locus containing IFITM5 and various numbers of IR-IFITM genes. Conservation of evolutionary synteny was observed in these IFITM genes. Significant functional divergence was detected among the three IFITM sub-families. No gene duplication or positive selection was found in IFITM5 sub-family, implying the functional conservation of IFITM5 in vertebrate evolution, which is involved in bone formation. No IFITM5 locus was identified in the marmoset genome, suggesting a potential association with the tiny size of this monkey. The IFITM10 sub-family was divided into two groups: aquatic and terrestrial types. Functional divergence was detected between the two groups, and five IFITM10-like genes from frog were dispersed into the two groups. Both gene duplication and positive selection were observed in aquatic vertebrate IFITM10-like genes, indicating that IFITM10 might be associated with the adaptation to aquatic environments. A large number of lineage- and species-specific gene duplications were observed in IR-IFITM sub-family and positive selection was detected in IR-IFITM of primates and rodents. Because primates have experienced a long history of viral infection, such rapid expansion and positive selection suggests that the evolution of primate IR-IFITM genes is associated with broad-spectrum antiviral activity.
Conflict of interest statement
Figures


Similar articles
-
Evolution of vertebrate interferon inducible transmembrane proteins.BMC Genomics. 2012 Apr 26;13:155. doi: 10.1186/1471-2164-13-155. BMC Genomics. 2012. PMID: 22537233 Free PMC article.
-
The vertebrate makorin ubiquitin ligase gene family has been shaped by large-scale duplication and retroposition from an ancestral gonad-specific, maternal-effect gene.BMC Genomics. 2010 Dec 20;11:721. doi: 10.1186/1471-2164-11-721. BMC Genomics. 2010. PMID: 21172006 Free PMC article.
-
The role of gene duplication and unconstrained selective pressures in the melanopsin gene family evolution and vertebrate circadian rhythm regulation.PLoS One. 2012;7(12):e52413. doi: 10.1371/journal.pone.0052413. Epub 2012 Dec 21. PLoS One. 2012. PMID: 23285031 Free PMC article.
-
Evolutionary dynamics of olfactory and other chemosensory receptor genes in vertebrates.J Hum Genet. 2006;51(6):505-517. doi: 10.1007/s10038-006-0391-8. Epub 2006 Apr 11. J Hum Genet. 2006. PMID: 16607462 Free PMC article. Review.
-
The small interferon-induced transmembrane genes and proteins.J Interferon Cytokine Res. 2011 Jan;31(1):183-97. doi: 10.1089/jir.2010.0112. Epub 2010 Dec 19. J Interferon Cytokine Res. 2011. PMID: 21166591 Review.
Cited by
-
Interferon-induced transmembrane protein 3 is a type II transmembrane protein.J Biol Chem. 2013 Nov 8;288(45):32184-32193. doi: 10.1074/jbc.M113.514356. Epub 2013 Sep 25. J Biol Chem. 2013. PMID: 24067232 Free PMC article.
-
Interferon-induced transmembrane protein 1 regulates endothelial lumen formation during angiogenesis.Arterioscler Thromb Vasc Biol. 2014 May;34(5):1011-9. doi: 10.1161/ATVBAHA.114.303352. Epub 2014 Mar 6. Arterioscler Thromb Vasc Biol. 2014. PMID: 24603679 Free PMC article.
-
Current Progress on Host Antiviral Factor IFITMs.Front Immunol. 2020 Nov 30;11:543444. doi: 10.3389/fimmu.2020.543444. eCollection 2020. Front Immunol. 2020. PMID: 33329509 Free PMC article. Review.
-
Evolution of primate interferon-induced transmembrane proteins (IFITMs): a story of gain and loss with a differentiation into a canonical cluster and IFITM retrogenes.Front Microbiol. 2023 Jul 26;14:1213685. doi: 10.3389/fmicb.2023.1213685. eCollection 2023. Front Microbiol. 2023. PMID: 37577422 Free PMC article.
-
Chicken interferon-inducible transmembrane protein 3 restricts influenza viruses and lyssaviruses in vitro.J Virol. 2013 Dec;87(23):12957-66. doi: 10.1128/JVI.01443-13. Epub 2013 Sep 25. J Virol. 2013. PMID: 24067955 Free PMC article.
References
-
- Chen YX, Welte K, Gebhard DH, Evans RL (1984) Induction of T cell aggregation by antibody to a 16kd human leukocyte surface antigen. J Immunol 133: 2496–2501. - PubMed
-
- Siegrist F, Ebeling M, Certa U (2011) The small interferon-induced transmembrane genes and proteins. J Interferon Cytokine Res 31: 183–197. - PubMed
-
- Moffatt P, Gaumond MH, Salois P, Sellin K, Bessette MC, et al. (2008) Bril: a novel bone-specific modulator of mineralization. J Bone Miner Res 23: 1497–1508. - PubMed
-
- Hanagata N, Li × (2011) Osteoblast-enriched membrane protein IFITM5 regulates the association of CD9 with an FKBP11-CD81-FPRP complex and stimulates expression of interferon-induced genes. Biochem Biophys Res Commun 409: 378–384. - PubMed
-
- Daniel-Carmi V, Makovitzki-Avraham E, Reuven EM, Goldstein I, Zilkha N, et al. (2009) The human 1–8D gene (IFITM2) is a novel p53 independent pro-apoptotic gene. Int J Cancer 125: 2810–2819. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

