Comparative analysis of genome sequences covering the seven cronobacter species
- PMID: 23166675
- PMCID: PMC3500316
- DOI: 10.1371/journal.pone.0049455
Comparative analysis of genome sequences covering the seven cronobacter species
Abstract
Background: Species of Cronobacter are widespread in the environment and are occasional food-borne pathogens associated with serious neonatal diseases, including bacteraemia, meningitis, and necrotising enterocolitis. The genus is composed of seven species: C. sakazakii, C. malonaticus, C. turicensis, C. dublinensis, C. muytjensii, C. universalis, and C. condimenti. Clinical cases are associated with three species, C. malonaticus, C. turicensis and, in particular, with C. sakazakii multilocus sequence type 4. Thus, it is plausible that virulence determinants have evolved in certain lineages.
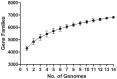
Methodology/principal findings: We generated high quality sequence drafts for eleven Cronobacter genomes representing the seven Cronobacter species, including an ST4 strain of C. sakazakii. Comparative analysis of these genomes together with the two publicly available genomes revealed Cronobacter has over 6,000 genes in one or more strains and over 2,000 genes shared by all Cronobacter. Considerable variation in the presence of traits such as type six secretion systems, metal resistance (tellurite, copper and silver), and adhesins were found. C. sakazakii is unique in the Cronobacter genus in encoding genes enabling the utilization of exogenous sialic acid which may have clinical significance. The C. sakazakii ST4 strain 701 contained additional genes as compared to other C. sakazakii but none of them were known specific virulence-related genes.
Conclusions/significance: Genome comparison revealed that pair-wise DNA sequence identity varies between 89 and 97% in the seven Cronobacter species, and also suggested various degrees of divergence. Sets of universal core genes and accessory genes unique to each strain were identified. These gene sequences can be used for designing genus/species specific detection assays. Genes encoding adhesins, T6SS, and metal resistance genes as well as prophages are found in only subsets of genomes and have contributed considerably to the variation of genomic content. Differences in gene content likely contribute to differences in the clinical and environmental distribution of species and sequence types.
Conflict of interest statement
Figures


References
-
- Friedemann M (2007) Enterobacter sakazakii in food and beverages (other than infant formula and milk powder). Int J Food Microbiol 116: 1–10. - PubMed
-
- Kandhai MC, Reij MW, Gorris LG, Guillaume-Gentil O, van Schothorst M (2004) Occurrence of Enterobacter sakazakii in food production environments and households. Lancet 363: 39–40. - PubMed
-
- Kucerova E, Joseph S, Forsythe SJ (2011) The Cronobacter genus: ubiquity and diversity. Quality Assurance and Safety of Crops & Foods 3: 104–122.
-
- FAO/WHO (2008) Enterobacter sakazakii (Cronobacter spp.) in powdered follow-up formulae. Microbiological Risk Assessment Series No. 15, Rome, 90pp. Available: http://www.who.int/foodsafety/publications/micro/mra_followup/en/.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

