An altered immune response, but not individual cationic antimicrobial peptides, is associated with the oral attenuation of Ara4N-deficient Salmonella enterica serovar Typhimurium in mice
- PMID: 23166721
- PMCID: PMC3499468
- DOI: 10.1371/journal.pone.0049588
An altered immune response, but not individual cationic antimicrobial peptides, is associated with the oral attenuation of Ara4N-deficient Salmonella enterica serovar Typhimurium in mice
Abstract
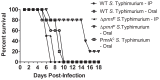
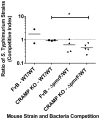
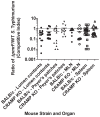
Salmonella enterica serovar Typhimurium (S. Typhimurium) uses two-component regulatory systems (TCRS) to respond to stimuli in the local microenvironment. Upon infection, the Salmonella TCRSs PhoP-PhoQ (PhoPQ) and PmrA-PmrB (PmrAB) are activated by environmental signals in the intestinal lumen and within host cells. TCRS-mediated gene expression results in lipopolysaccharide (LPS) modification and cationic antimicrobial peptide resistance. The PmrA-regulated pmrHFIJKLM operon mediates 4-amino-4-deoxy-L-arabinose (Ara4N) production and attachment to the lipid A of LPS. A ΔpmrF S. Typhimurium strain cannot produce Ara4N, exhibits increased sensitivity to cationic antimicrobial peptide (CAMP)-mediated killing, and attenuated virulence in mice upon oral infection. CAMPs are predicted to play a role in elimination of Salmonella, and may activate PhoPQ and PmrAB in vivo, which could increase bacterial resistance to host defenses. Competition experiments between wild type (WT) and ΔpmrF mutant strains of S. Typhimurium indicated that selection against this mutant first occurs within the intestinal lumen early during infection. However, CRAMP and active cryptdins alone are not responsible for elimination of Ara4N-deficient bacteria in vivo. Investigation into the early immune response to ΔpmrF showed that it differed slightly from the early immune response to WT S. Typhimurium. Further investigation into the early immune response to infection of Peyer's patches suggests a role for IL-13 in the attenution of the ΔpmrF mutant strain. Thus, prominent CAMPs present in the mouse intestine are not responsible for the selection against the ΔpmrF strain in this location, but limited alterations in innate immune induction were observed that affect bacterial survival and virulence.
Conflict of interest statement
Figures
Similar articles
-
Cationic antimicrobial peptides serve as activation signals for the Salmonella Typhimurium PhoPQ and PmrAB regulons in vitro and in vivo.Front Cell Infect Microbiol. 2012 Jul 27;2:102. doi: 10.3389/fcimb.2012.00102. eCollection 2012. Front Cell Infect Microbiol. 2012. PMID: 22919691 Free PMC article.
-
Genetic and functional analysis of a PmrA-PmrB-regulated locus necessary for lipopolysaccharide modification, antimicrobial peptide resistance, and oral virulence of Salmonella enterica serovar typhimurium.Infect Immun. 2000 Nov;68(11):6139-46. doi: 10.1128/IAI.68.11.6139-6146.2000. Infect Immun. 2000. PMID: 11035717 Free PMC article.
-
Cathelicidin antimicrobial peptide expression is not induced or required for bacterial clearance during salmonella enterica infection of human monocyte-derived macrophages.Infect Immun. 2012 Nov;80(11):3930-8. doi: 10.1128/IAI.00672-12. Epub 2012 Aug 27. Infect Immun. 2012. PMID: 22927052 Free PMC article.
-
The Salmonella PmrAB regulon: lipopolysaccharide modifications, antimicrobial peptide resistance and more.Trends Microbiol. 2008 Jun;16(6):284-90. doi: 10.1016/j.tim.2008.03.007. Epub 2008 May 6. Trends Microbiol. 2008. PMID: 18467098 Review.
-
S. Typhimurium strategies to resist killing by cationic antimicrobial peptides.Biochim Biophys Acta. 2015 Nov;1848(11 Pt B):3021-5. doi: 10.1016/j.bbamem.2015.01.013. Epub 2015 Jan 30. Biochim Biophys Acta. 2015. PMID: 25644871 Free PMC article. Review.
Cited by
-
Antibiotic development challenges: the various mechanisms of action of antimicrobial peptides and of bacterial resistance.Front Microbiol. 2013 Dec 9;4:353. doi: 10.3389/fmicb.2013.00353. Front Microbiol. 2013. PMID: 24367355 Free PMC article. Review.
-
Characterization of Canine Peyer's Patches by Multidimensional Analysis: Insights from Immunofluorescence, Flow Cytometry, and Single-Cell RNA Sequencing.Immunohorizons. 2023 Nov 1;7(11):788-805. doi: 10.4049/immunohorizons.2300091. Immunohorizons. 2023. PMID: 38015460 Free PMC article.
-
Mechanisms of Antimicrobial Peptide Resistance in Gram-Negative Bacteria.Antibiotics (Basel). 2015 Mar;4(1):18-41. doi: 10.3390/antibiotics4010018. Antibiotics (Basel). 2015. PMID: 25927010 Free PMC article.
-
Ail and PagC-related proteins in the entomopathogenic bacteria of Photorhabdus genus.PLoS One. 2014 Oct 15;9(10):e110060. doi: 10.1371/journal.pone.0110060. eCollection 2014. PLoS One. 2014. PMID: 25333642 Free PMC article.
-
Antimicrobial peptide exposure selects for Staphylococcus aureus resistance to human defence peptides.J Antimicrob Chemother. 2017 Jan;72(1):115-127. doi: 10.1093/jac/dkw381. Epub 2016 Sep 20. J Antimicrob Chemother. 2017. PMID: 27650186 Free PMC article.
References
-
- Jones DE, Bevins CL (1992) Paneth cells of the human small intestine express an antimicrobial peptide gene. J Biol Chem 267: 23216–23225. - PubMed
-
- Zasloff M (1992) Antibiotic peptides as mediators of innate immunity. Curr Opin Immunol 4: 3–7. - PubMed
-
- Radek K, Gallo R (2007) Antimicrobial peptides: natural effectors of the innate immune system. Semin Immunopathol 29: 27–43. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

