Use of four next-generation sequencing platforms to determine HIV-1 coreceptor tropism
- PMID: 23166726
- PMCID: PMC3498215
- DOI: 10.1371/journal.pone.0049602
Use of four next-generation sequencing platforms to determine HIV-1 coreceptor tropism
Abstract
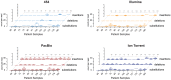
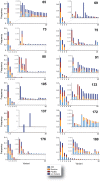
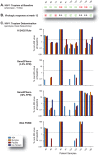
HIV-1 coreceptor tropism assays are required to rule out the presence of CXCR4-tropic (non-R5) viruses prior treatment with CCR5 antagonists. Phenotypic (e.g., Trofile™, Monogram Biosciences) and genotypic (e.g., population sequencing linked to bioinformatic algorithms) assays are the most widely used. Although several next-generation sequencing (NGS) platforms are available, to date all published deep sequencing HIV-1 tropism studies have used the 454™ Life Sciences/Roche platform. In this study, HIV-1 co-receptor usage was predicted for twelve patients scheduled to start a maraviroc-based antiretroviral regimen. The V3 region of the HIV-1 env gene was sequenced using four NGS platforms: 454™, PacBio® RS (Pacific Biosciences), Illumina®, and Ion Torrent™ (Life Technologies). Cross-platform variation was evaluated, including number of reads, read length and error rates. HIV-1 tropism was inferred using Geno2Pheno, Web PSSM, and the 11/24/25 rule and compared with Trofile™ and virologic response to antiretroviral therapy. Error rates related to insertions/deletions (indels) and nucleotide substitutions introduced by the four NGS platforms were low compared to the actual HIV-1 sequence variation. Each platform detected all major virus variants within the HIV-1 population with similar frequencies. Identification of non-R5 viruses was comparable among the four platforms, with minor differences attributable to the algorithms used to infer HIV-1 tropism. All NGS platforms showed similar concordance with virologic response to the maraviroc-based regimen (75% to 80% range depending on the algorithm used), compared to Trofile (80%) and population sequencing (70%). In conclusion, all four NGS platforms were able to detect minority non-R5 variants at comparable levels suggesting that any NGS-based method can be used to predict HIV-1 coreceptor usage.
Conflict of interest statement
Figures




References
-
- Dragic T, Litwin V, Allaway GP, Martin SR, Huang Y, et al. (1996) HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC- CKR-5. Nature 381: 667–673. - PubMed
-
- Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, et al. (1996) Identification of a major co-receptor for primary isolates of HIV-1. Nature 381: 661–666. - PubMed
-
- Tilton JC, Doms RW (2010) Entry inhibitors in the treatment of HIV-1 infection. Antiviral research 85: 91–100. - PubMed
-
- Dorr P, Westby M, Dobbs S, Griffin P, Irvine B, et al. (2005) Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. AntimicrobAgents Chemother 49: 4721–4732. - PMC - PubMed
-
- Westby M, Lewis M, Whitcomb J, Youle M, Pozniak AL, et al. (2006) Emergence of CXCR4-using human immunodeficiency virus type 1 (HIV-1) variants in a minority of HIV-1-infected patients following treatment with the CCR5 antagonist maraviroc is from a pretreatment CXCR4-using virus reservoir. Journal of Virology 80: 4909–4920. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

