TMV-Gate vectors: gateway compatible tobacco mosaic virus based expression vectors for functional analysis of proteins
- PMID: 23166857
- PMCID: PMC3500846
- DOI: 10.1038/srep00874
TMV-Gate vectors: gateway compatible tobacco mosaic virus based expression vectors for functional analysis of proteins
Abstract
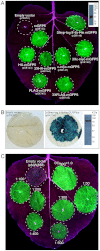
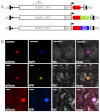
Plant viral expression vectors are advantageous for high-throughput functional characterization studies of genes due to their capability for rapid, high-level transient expression of proteins. We have constructed a series of tobacco mosaic virus (TMV) based vectors that are compatible with Gateway technology to enable rapid assembly of expression constructs and exploitation of ORFeome collections. In addition to the potential of producing recombinant protein at grams per kilogram FW of leaf tissue, these vectors facilitate either N- or C-terminal fusions to a broad series of epitope tag(s) and fluorescent proteins. We demonstrate the utility of these vectors in affinity purification, immunodetection and subcellular localisation studies. We also apply the vectors to characterize protein-protein interactions and demonstrate their utility in screening plant pathogen effectors. Given its broad utility in defining protein properties, this vector series will serve as a useful resource to expedite gene characterization efforts.
Figures





Similar articles
-
TRBO: a high-efficiency tobacco mosaic virus RNA-based overexpression vector.Plant Physiol. 2007 Dec;145(4):1232-40. doi: 10.1104/pp.107.106377. Epub 2007 Aug 24. Plant Physiol. 2007. PMID: 17720752 Free PMC article.
-
High-efficiency protein expression in plants from agroinfection-compatible Tobacco mosaic virus expression vectors.BMC Biotechnol. 2007 Aug 27;7:52. doi: 10.1186/1472-6750-7-52. BMC Biotechnol. 2007. PMID: 17723150 Free PMC article.
-
Potatovirus X and Tobacco mosaic virus-based vectors compatible with the Gateway cloning system.J Virol Methods. 2010 Mar;164(1-2):7-13. doi: 10.1016/j.jviromet.2009.11.005. Epub 2009 Nov 10. J Virol Methods. 2010. PMID: 19903495
-
Plant virus expression vectors set the stage as production platforms for biopharmaceutical proteins.Virology. 2012 Nov 10;433(1):1-6. doi: 10.1016/j.virol.2012.06.012. Virology. 2012. PMID: 22979981 Review.
-
Recent Developments in Design and Application of Plant Virus-based Gene Vectors.Recent Pat DNA Gene Seq. 2007;1(3):214-26. doi: 10.2174/187221507782360218. Recent Pat DNA Gene Seq. 2007. PMID: 19075936 Review.
Cited by
-
Recent advances on host plants and expression cassettes' structure and function in plant molecular pharming.BioDrugs. 2014 Apr;28(2):145-59. doi: 10.1007/s40259-013-0062-1. BioDrugs. 2014. PMID: 23959796 Free PMC article. Review.
-
Production of Bioactive Recombinant Reteplase by Virus-Based Transient Expression System in Nicotiana benthamiana.Front Plant Sci. 2019 Oct 8;10:1225. doi: 10.3389/fpls.2019.01225. eCollection 2019. Front Plant Sci. 2019. PMID: 31649696 Free PMC article.
-
Transient Production of Human β-Glucocerebrosidase With Mannosidic-Type N-Glycan Structure in Glycoengineered Nicotiana benthamiana Plants.Front Plant Sci. 2021 Jun 7;12:683762. doi: 10.3389/fpls.2021.683762. eCollection 2021. Front Plant Sci. 2021. PMID: 34163514 Free PMC article.
-
Exploiting plant virus-derived components to achieve in planta expression and for templates for synthetic biology applications.New Phytol. 2013 Oct;200(1):16-26. doi: 10.1111/nph.12204. Epub 2013 Mar 4. New Phytol. 2013. PMID: 23452220 Free PMC article. Review.
-
Transient Expression of Tetrameric Recombinant Human Butyrylcholinesterase in Nicotiana benthamiana.Front Plant Sci. 2016 Jun 16;7:743. doi: 10.3389/fpls.2016.00743. eCollection 2016. Front Plant Sci. 2016. PMID: 27379103 Free PMC article.
References
-
- Scholthof H. B., Scholthof K. B. & Jackson A. O. Plant virus gene vectors for transient expression of foreign proteins in plants. Annu Rev Phytopathol 34, 299–323 (1996). - PubMed
-
- Yusibov V., Shivprasad S., Turpen T. H., Dawson W. & Koprowski H. Plant viral vectors based on tobamoviruses. Curr Top Microbiol Immunol 240, 81–94 (1999). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

