Whole exome sequencing in a patient with uniparental disomy of chromosome 2 and a complex phenotype
- PMID: 23167750
- PMCID: PMC3996682
- DOI: 10.1111/cge.12064
Whole exome sequencing in a patient with uniparental disomy of chromosome 2 and a complex phenotype
Abstract
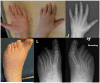
Whole exome sequencing and chromosomal microarrays are two powerful technologies that have transformed the ability of researchers to search for potentially causal variants in human disease. This study combines these tools to search for causal variants in a patient found to have maternal uniparental isodisomy of chromosome 2. This subject has a complex phenotype including skeletal and renal dysplasia, immune deficiencies, growth failure, retinal degeneration and ovarian insufficiency. Eighteen non-synonymous, rare homozygous variants were identified on chromosome 2. Additionally, five genes with compound heterozygous mutations were detected on other chromosomes that could lead to a disease phenotype independent of the uniparental disomy found in this case. Several candidate genes with potential connection to the phenotype are described but none are definitively proven to be causal. This study highlights the potential for detection of a large number of candidate genes using whole exome sequencing complicating interpretation in both the research and clinical settings. Forums must be created for publication and sharing of detailed phenotypic and genotypic reports to facilitate further biological discoveries and clinical counseling.
Keywords: Bardet-Biedl syndrome; DNA copy number variations; comparative genomic hybridization; high-throughput nucleotide sequencing; uniparental disomy; whole exome sequencing.
© 2012 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd.
Figures


References
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

