Periplasmic production via the pET expression system of soluble, bioactive human growth hormone
- PMID: 23168094
- PMCID: PMC3537859
- DOI: 10.1016/j.pep.2012.11.002
Periplasmic production via the pET expression system of soluble, bioactive human growth hormone
Abstract
A pET based expression system for the production of recombinant human growth hormone (hGH) directed to the Escherichia coli periplasmic space was developed. The pET22b plasmid was used as a template for creating vectors that encode hGH fused to either a pelB or ompA secretion signal under control of the strong bacteriophage T7 promoter. The pelB- and ompA-hGH constructs expressed in BL21 (λDE3)-RIPL E. coli are secreted into the periplasm which facilitates isolation of soluble hGH by selective disruption of the outer membrane. A carboxy-terminal poly-histidine tag enabled purification by Ni(2+) affinity chromatography with an average yield of 1.4 mg/L culture of purified hGH, independent of secretion signal. Purified pelB- and ompA-hGH are monomeric based on size exclusion chromatography with an intact mass corresponding to mature hGH indicating proper cleavage of the signal peptide and folding in the periplasm. Both pelB- and ompA-hGH bind the hGH receptor with high affinity and potently stimulate Nb2 cell growth. These results demonstrate that the pET expression system is suitable for the rapid and simple isolation of bioactive, soluble hGH from E. coli.
Copyright © 2012 Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures


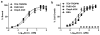

Similar articles
-
Engineered small metal-binding protein tag improves the production of recombinant human growth hormone in the periplasm of Escherichia coli.FEBS Open Bio. 2020 Apr;10(4):546-551. doi: 10.1002/2211-5463.12808. Epub 2020 Mar 9. FEBS Open Bio. 2020. PMID: 32049439 Free PMC article.
-
Soluble Expression, One-Step Purification and Characterization of Recombinant Human Growth Hormone Fused with ompA3 in Escherichia coli.Protein Pept Lett. 2021;28(5):533-542. doi: 10.2174/0929866527666201110123426. Protein Pept Lett. 2021. PMID: 33172365
-
Expression, purification and characterization of the authentic form of human growth hormone receptor antagonist G120R-hGH obtained in Escherichia coli periplasmic space.Protein Expr Purif. 2017 Mar;131:91-100. doi: 10.1016/j.pep.2016.12.001. Epub 2016 Dec 22. Protein Expr Purif. 2017. PMID: 28013084
-
Escherichia coli "TatExpress" strains export several g/L human growth hormone to the periplasm by the Tat pathway.Biotechnol Bioeng. 2019 Dec;116(12):3282-3291. doi: 10.1002/bit.27147. Epub 2019 Sep 2. Biotechnol Bioeng. 2019. PMID: 31429928 Free PMC article.
-
Purification of secreted recombinant proteins from Escherichia coli.Bioprocess Technol. 1991;12:163-81. Bioprocess Technol. 1991. PMID: 1367083 Review.
Cited by
-
Periplasmic Expression of TNF Related Apoptosis Inducing Ligand (TRAIL) in E.coli.Iran J Pharm Res. 2015 Spring;14(2):617-26. Iran J Pharm Res. 2015. PMID: 25901171 Free PMC article.
-
Twin arginine translocation system in secretory expression of recombinant human growth hormone.Res Pharm Sci. 2016 Dec;11(6):461-469. doi: 10.4103/1735-5362.194871. Res Pharm Sci. 2016. PMID: 28003839 Free PMC article.
-
Short stature explained by dimerization of human growth hormone induced by a p.C53S point mutation.J Biol Chem. 2020 Apr 10;295(15):4893-4901. doi: 10.1074/jbc.RA119.009101. Epub 2020 Mar 4. J Biol Chem. 2020. PMID: 32132170 Free PMC article.
-
Evaluation of Recombinant Human Growth Hormone Secretion in E. coli using the L-asparaginase II Signal Peptide.Avicenna J Med Biotechnol. 2016 Oct-Dec;8(4):182-187. Avicenna J Med Biotechnol. 2016. PMID: 27920886 Free PMC article.
-
A Novel Cadherin-like Protein Mediates Adherence to and Killing of Host Cells by the Parasite Trichomonas vaginalis.mBio. 2019 May 14;10(3):e00720-19. doi: 10.1128/mBio.00720-19. mBio. 2019. PMID: 31088924 Free PMC article.
References
-
- Itakura K, Hirose T, Crea R, Riggs AD, Heyneker HL, Bolivar F, Boyer HW. Expression in Escherichia coli of a chemically synthesized gene for the hormone somatostatin. Science. 1977;198:1056–63. - PubMed
-
- Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–30. - PubMed
-
- Yoon SH, Kim SK, Kim JF. Secretory production of recombinant proteins in Escherichia coli. Recent Pat Biotechnol. 2010;4:23–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

