Chemogenomic approach identified yeast YLR143W as diphthamide synthetase
- PMID: 23169644
- PMCID: PMC3523822
- DOI: 10.1073/pnas.1214346109
Chemogenomic approach identified yeast YLR143W as diphthamide synthetase
Abstract
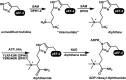
Many genes are of unknown functions in any sequenced genome. A combination of chemical and genetic perturbations has been used to investigate gene functions. Here we present a case that such "chemogenomics" information can be effectively used to identify missing genes in a defined biological pathway. In particular, we identified the previously unknown enzyme diphthamide synthetase for the last step of diphthamide biosynthesis. We found that yeast protein YLR143W is the diphthamide synthetase catalyzing the last amidation step using ammonium and ATP. Diphthamide synthetase is evolutionarily conserved in eukaryotes. The previously uncharacterized human gene ATPBD4 is the ortholog of yeast YLR143W and fully rescues the deletion of YLR143W in yeast.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Robinson EA, Henriksen O, Maxwell ES. Elongation factor 2. Amino acid sequence at the site of adenosine diphosphate ribosylation. J Biol Chem. 1974;249(16):5088–5093. - PubMed
-
- Van Ness BG, Howard JB, Bodley JW. ADP-ribosylation of elongation factor 2 by diphtheria toxin. NMR spectra and proposed structures of ribosyl-diphthamide and its hydrolysis products. J Biol Chem. 1980;255(22):10710–10716. - PubMed
-
- Van Ness BG, Howard JB, Bodley JW. ADP-ribosylation of elongation factor 2 by diphtheria toxin. Isolation and properties of the novel ribosyl-amino acid and its hydrolysis products. J Biol Chem. 1980;255(22):10717–10720. - PubMed
-
- Collier RJ. Understanding the mode of action of diphtheria toxin: A perspective on progress during the 20th century. Toxicon. 2001;39(11):1793–1803. - PubMed
-
- Moehring JM, Moehring TJ. The post-translational trimethylation of diphthamide studied in vitro. J Biol Chem. 1988;263(8):3840–3844. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

