Mutual protection of ribosomal proteins L5 and L11 from degradation is essential for p53 activation upon ribosomal biogenesis stress
- PMID: 23169665
- PMCID: PMC3528581
- DOI: 10.1073/pnas.1218535109
Mutual protection of ribosomal proteins L5 and L11 from degradation is essential for p53 activation upon ribosomal biogenesis stress
Abstract
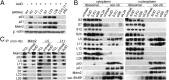
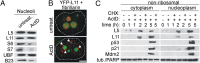
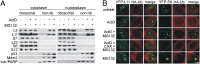
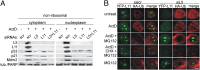
Impairment of ribosomal biogenesis can activate the p53 protein independently of DNA damage. The ability of ribosomal proteins L5, L11, L23, L26, or S7 to bind Mdm2 and inhibit its ubiquitin ligase activity has been suggested as a critical step in p53 activation under these conditions. Here, we report that L5 and L11 are particularly important for this response. Whereas several other newly synthesized ribosomal proteins are degraded by proteasomes upon inhibition of Pol I activity by actinomycin D, L5 and L11 accumulate in the ribosome-free fraction where they bind to Mdm2. This selective accumulation of free L5 and L11 is due to their mutual protection from proteasomal degradation. Furthermore, the endogenous, newly synthesized L5 and L11 continue to be imported into nucleoli even after nucleolar disruption and colocalize with Mdm2, p53, and promyelocytic leukemia protein. This suggests that the disrupted nucleoli may provide a platform for L5- and L11-dependent p53 activation, implying a role for the nucleolus in p53 activation by ribosomal biogenesis stress. These findings may have important implications with respect to understanding the pathogenesis of diseases caused by impaired ribosome biogenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

