Riboswitches for intracellular study of genes involved in Francisella pathogenesis
- PMID: 23169998
- PMCID: PMC3509410
- DOI: 10.1128/mBio.00253-12
Riboswitches for intracellular study of genes involved in Francisella pathogenesis
Abstract
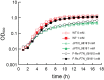
The study of many important intracellular bacterial pathogens requires an understanding of how specific virulence factors contribute to pathogenesis during the infection of host cells. This requires tools to dissect gene function, but unfortunately, there is a lack of such tools for research on many difficult-to-study, or understudied, intracellular pathogens. Riboswitches are RNA-based genetic control elements that directly modulate gene expression upon ligand binding. Here we report the application of theophylline-sensitive synthetic riboswitches to induce protein expression in the intracellular pathogen Francisella. We show that this system can be used to activate the bacterial expression of the reporter β-galactosidase during growth in rich medium. Furthermore, we applied this system to control the expression of green fluorescent protein during intracellular infection by the addition of theophylline directly to infected macrophages. Importantly, we could control the expression of a novel endogenous protein required for growth under nutrient-limiting conditions and replication in macrophages, FTN_0818. Riboswitch-mediated control of FTN_0818 rescued the growth of an FTN_0818 mutant in minimal medium and during macrophage infection. This is the first demonstration of the use of a synthetic riboswitch to control an endogenous gene required for a virulence trait in an intracellular bacterium. Since this system can be adapted to diverse bacteria, the ability to use riboswitches to regulate intracellular bacterial gene expression will likely facilitate the in-depth study of the virulence mechanisms of numerous difficult-to-study intracellular pathogens such as Ehrlichia chaffeensis, Anaplasma phagocytophilum, and Orientia tsutsugamushi, as well as future emerging pathogens.
Importance: Determining how specific bacterial genes contribute to virulence during the infection of host cells is critical to understanding how pathogens cause disease. This can be especially challenging with many difficult-to-study intracellular pathogens. Riboswitches are RNA-based genetic control elements that can be used to help dissect gene function, especially since they can be used in a broad range of bacteria. We demonstrate the utility of riboswitches, and for the first time show that riboswitches can be used to functionally control a bacterial gene that is critical to the ability of a pathogen to cause disease, during intracellular infection. Since this system can be adapted to diverse bacteria, riboswitches will likely facilitate the in-depth study of the virulence mechanisms of numerous difficult-to-study intracellular pathogens, as well as future emerging pathogens.
Figures




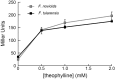
References
-
- Sørensen HP, Mortensen KK. 2005. Advanced genetic strategies for recombinant protein expression in Escherichia coli. J. Biotechnol. 115:113–128 - PubMed
-
- Baneyx F. 1999. Recombinant protein expression in Escherichia coli. Curr. Opin. Biotechnol. 10:411–421 - PubMed
-
- Scaife J, Leach DRF, Galizzi A. 1985. Genetics of bacteria. Academic Press, New York, NY.
-
- Winkler WC, Breaker RR. 2005. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 59:487–517 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
