The membrane-proximal region (MPR) of herpes simplex virus gB regulates association of the fusion loops with lipid membranes
- PMID: 23170000
- PMCID: PMC3509434
- DOI: 10.1128/mBio.00429-12
The membrane-proximal region (MPR) of herpes simplex virus gB regulates association of the fusion loops with lipid membranes
Abstract
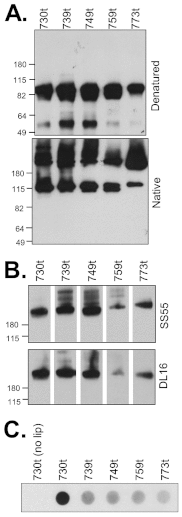
Glycoprotein B (gB), gD, and gH/gL constitute the fusion machinery of herpes simplex virus (HSV). Prior studies indicated that fusion occurs in a stepwise fashion whereby the gD/receptor complex activates the entire process, while gH/gL regulates the fusion reaction carried out by gB. Trimeric gB is a class III fusion protein. Its ectodomain of 773 amino acids contains a membrane-proximal region (MPR) (residues 731 to 773) and two fusion loops (FLs) per protomer. We hypothesized that the highly hydrophobic MPR interacts with the FLs, thereby masking them on virions until fusion begins. To test this hypothesis, we made a series of deletion, truncation, and point mutants of the gB MPR. Although the full-length deletion mutants were expressed in transfected cells, they were not transported to the cell surface, suggesting that removal of even small stretches of the MPR was highly detrimental to gB folding. To circumvent this limitation, we used a baculovirus expression system to generate four soluble proteins, each lacking the transmembrane region and cytoplasmic tail. All retained the FLs and decreasing portions of the MPR [gB(773t) (gB truncated at amino acid 773), gB(759t), gB(749t), and gB(739t)]. Despite the presence of the FLs, all were compromised in their ability to bind liposomes compared to the control, gB(730t), which lacks the MPR. We conclude that residues 731 to 739 are sufficient to mask the FLs, thereby preventing liposome association. Importantly, mutation of two aromatic residues (F732 and F738) to alanine restored the ability of gB(739t) to bind liposomes. Our data suggest that the MPR is important for modulating the association of gB FLs with target membranes. IMPORTANCE To successfully cause disease, a virus must infect host cells. Viral infection is a highly regulated, multistep process. For herpesviruses, genetic material transfers from the virus to the target cell through fusion of the viral and host cell lipid membranes. Here, we provide evidence that the ability of the herpes simplex virus (HSV) glycoprotein B (gB) fusion protein to interact with the host membrane is regulated by its membrane-proximal region (MPR), which serves to cover or shield its lipid-associating moieties (fusion loops). This in turn prevents the premature binding of gB with host cells and provides a level of regulation to the fusion process. These findings provide important insight into the complex regulatory steps required for successful herpesvirus infection.
Figures


Similar articles
-
Herpes simplex virus glycoprotein B associates with target membranes via its fusion loops.J Virol. 2009 Jul;83(13):6825-36. doi: 10.1128/JVI.00301-09. Epub 2009 Apr 15. J Virol. 2009. PMID: 19369321 Free PMC article.
-
Functional Characterization of Glycoprotein H Chimeras Composed of Conserved Domains of the Pseudorabies Virus and Herpes Simplex Virus 1 Homologs.J Virol. 2015 Oct 21;90(1):421-32. doi: 10.1128/JVI.01985-15. Print 2016 Jan 1. J Virol. 2015. PMID: 26491153 Free PMC article.
-
Multiple Sites on Glycoprotein H (gH) Functionally Interact with the gB Fusion Protein to Promote Fusion during Herpes Simplex Virus (HSV) Entry.mBio. 2023 Feb 28;14(1):e0336822. doi: 10.1128/mbio.03368-22. Epub 2023 Jan 11. mBio. 2023. PMID: 36629412 Free PMC article.
-
Two Sides to Every Story: Herpes Simplex Type-1 Viral Glycoproteins gB, gD, gH/gL, gK, and Cellular Receptors Function as Key Players in Membrane Fusion.Viruses. 2021 Sep 16;13(9):1849. doi: 10.3390/v13091849. Viruses. 2021. PMID: 34578430 Free PMC article. Review.
-
Herpesvirus membrane fusion - a team effort.Curr Opin Struct Biol. 2020 Jun;62:112-120. doi: 10.1016/j.sbi.2019.12.004. Epub 2020 Jan 11. Curr Opin Struct Biol. 2020. PMID: 31935542 Review.
Cited by
-
The Fusion Loops of the Initial Prefusion Conformation of Herpes Simplex Virus 1 Fusion Protein Point Toward the Membrane.mBio. 2017 Aug 22;8(4):e01268-17. doi: 10.1128/mBio.01268-17. mBio. 2017. PMID: 28830949 Free PMC article.
-
Membrane fusion, potential threats, and natural antiviral drugs of pseudorabies virus.Vet Res. 2023 May 2;54(1):39. doi: 10.1186/s13567-023-01171-z. Vet Res. 2023. PMID: 37131259 Free PMC article. Review.
-
Herpes Simplex Virus Cell Entry Mechanisms: An Update.Front Cell Infect Microbiol. 2021 Jan 18;10:617578. doi: 10.3389/fcimb.2020.617578. eCollection 2020. Front Cell Infect Microbiol. 2021. PMID: 33537244 Free PMC article. Review.
-
Cell entry mechanisms of HSV: what we have learned in recent years.Future Virol. 2015 Oct 1;10(10):1145-1154. doi: 10.2217/fvl.15.85. Future Virol. 2015. PMID: 27066105 Free PMC article.
-
Different functional states of fusion protein gB revealed on human cytomegalovirus by cryo electron tomography with Volta phase plate.PLoS Pathog. 2018 Dec 3;14(12):e1007452. doi: 10.1371/journal.ppat.1007452. eCollection 2018 Dec. PLoS Pathog. 2018. PMID: 30507948 Free PMC article.
References
-
- Browne H, Bruun B, Minson T. 2001. Plasma membrane requirements for cell fusion induced by herpes simplex virus type 1 glycoproteins gB, gD, gH and gL. J. Gen. Virol. 82:1419–1422 - PubMed
-
- Pertel PE, Fridberg A, Parish ML, Spear PG. 2001. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology 279:313–324 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

