Evasion of influenza A viruses from innate and adaptive immune responses
- PMID: 23170167
- PMCID: PMC3499814
- DOI: 10.3390/v4091438
Evasion of influenza A viruses from innate and adaptive immune responses
Abstract
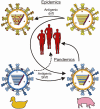
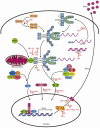
The influenza A virus is one of the leading causes of respiratory tract infections in humans. Upon infection with an influenza A virus, both innate and adaptive immune responses are induced. Here we discuss various strategies used by influenza A viruses to evade innate immune responses and recognition by components of the humoral and cellular immune response, which consequently may result in reduced clearing of the virus and virus-infected cells. Finally, we discuss how the current knowledge about immune evasion can be used to improve influenza A vaccination strategies.
Keywords: adaptive immunity; evasion; influenza; innate immunity.
Figures
Similar articles
-
Modulation of Innate Immune Responses by the Influenza A NS1 and PA-X Proteins.Viruses. 2018 Dec 12;10(12):708. doi: 10.3390/v10120708. Viruses. 2018. PMID: 30545063 Free PMC article. Review.
-
Innate Immune Responses to Influenza Virus Infections in the Upper Respiratory Tract.Viruses. 2021 Oct 17;13(10):2090. doi: 10.3390/v13102090. Viruses. 2021. PMID: 34696520 Free PMC article. Review.
-
Identification of lncRNA-155 encoded by MIR155HG as a novel regulator of innate immunity against influenza A virus infection.Cell Microbiol. 2019 Aug;21(8):e13036. doi: 10.1111/cmi.13036. Epub 2019 May 29. Cell Microbiol. 2019. PMID: 31045320
-
Cell type- and replication stage-specific influenza virus responses in vivo.PLoS Pathog. 2020 Aug 13;16(8):e1008760. doi: 10.1371/journal.ppat.1008760. eCollection 2020 Aug. PLoS Pathog. 2020. PMID: 32790753 Free PMC article.
-
Dendritic cells and influenza A virus infection.Virulence. 2012 Nov 15;3(7):603-8. doi: 10.4161/viru.21864. Epub 2012 Oct 17. Virulence. 2012. PMID: 23076333 Free PMC article. Review.
Cited by
-
Inflammation, immunity and potential target therapy of SARS-COV-2: A total scale analysis review.Food Chem Toxicol. 2021 Apr;150:112087. doi: 10.1016/j.fct.2021.112087. Epub 2021 Feb 25. Food Chem Toxicol. 2021. PMID: 33640537 Free PMC article. Review.
-
Long-term adaptation of the influenza A virus by escaping cytotoxic T-cell recognition.Sci Rep. 2016 Sep 15;6:33334. doi: 10.1038/srep33334. Sci Rep. 2016. PMID: 27629812 Free PMC article.
-
Infection Dynamics of Swine Influenza Virus in a Danish Pig Herd Reveals Recurrent Infections with Different Variants of the H1N2 Swine Influenza A Virus Subtype.Viruses. 2020 Sep 10;12(9):1013. doi: 10.3390/v12091013. Viruses. 2020. PMID: 32927910 Free PMC article.
-
Emerging Role of Mucosal Vaccine in Preventing Infection with Avian Influenza A Viruses.Viruses. 2020 Aug 7;12(8):862. doi: 10.3390/v12080862. Viruses. 2020. PMID: 32784697 Free PMC article. Review.
-
Molecular Docking and In-Silico Analysis of Natural Biomolecules against Dengue, Ebola, Zika, SARS-CoV-2 Variants of Concern and Monkeypox Virus.Int J Mol Sci. 2022 Sep 22;23(19):11131. doi: 10.3390/ijms231911131. Int J Mol Sci. 2022. PMID: 36232431 Free PMC article.
References
-
- Palese P., Shaw M.L. Fields Virology. 5th. Vol. 2. Lippincott Williams & Wilkins, a Wolters Kluwer Business; Philadelphia, PA, USA: 2007. Orthomyxoviridae: The viruses and their replication; pp. 1647–1689.
-
- WHO. Influenza (seasonal) fact sheet No 211. [(accessed on 31 March 2012)]. Available online: http://www.who.int/mediacentre/factsheets/fs211/en/index.html.
-
- Jagger B.W., Wise H.M., Kash J.C., Walters K.A., Wills N.M., Xiao Y.L., Dunfee R.L., Schwartzman L.M., Ozinsky A., Bell G.L., et al. An overlapping protein-coding region in influenza a virus segment 3 modulates the host response. Science. 2012;337:199–204. doi: 10.1126/science.1222213. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

