The response regulator SypE controls biofilm formation and colonization through phosphorylation of the syp-encoded regulator SypA in Vibrio fischeri
- PMID: 23171087
- PMCID: PMC3556205
- DOI: 10.1111/mmi.12109
The response regulator SypE controls biofilm formation and colonization through phosphorylation of the syp-encoded regulator SypA in Vibrio fischeri
Abstract
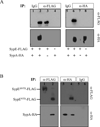
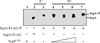
Bacteria utilize multiple regulatory systems to modulate gene expression in response to environmental changes, including two-component signalling systems and partner-switching networks. We recently identified a novel regulatory protein, SypE, that combines features of both signalling systems. SypE contains a central response regulator receiver domain flanked by putative kinase and phosphatase effector domains with similarity to partner-switching proteins. SypE was previously shown to exert dual control over biofilm formation through the opposing activities of its terminal effector domains. Here, we demonstrate that SypE controls biofilms in Vibrio fischeri by regulating the activity of SypA, a STAS (sulphate transporter and anti-sigma antagonist) domain protein. Using biochemical and genetic approaches, we determined that SypE both phosphorylates and dephosphorylates SypA, and that phosphorylation inhibits SypA's activity. Furthermore, we found that biofilm formation and symbiotic colonization required active, unphosphorylated SypA, and thus SypA phosphorylation corresponded with a loss of biofilms and impaired host colonization. Finally, expression of a non-phosphorylatable mutant of SypA suppressed both the biofilm and symbiosis defects of a constitutively inhibitory SypE mutant strain. This study demonstrates that regulation of SypA activity by SypE is a critical mechanism by which V. fischeri controls biofilm development and symbiotic colonization.
© 2012 Blackwell Publishing Ltd.
Figures








Similar articles
-
Inhibition of SypG-induced biofilms and host colonization by the negative regulator SypE in Vibrio fischeri.PLoS One. 2013;8(3):e60076. doi: 10.1371/journal.pone.0060076. Epub 2013 Mar 28. PLoS One. 2013. PMID: 23555890 Free PMC article.
-
Inactivation of a novel response regulator is necessary for biofilm formation and host colonization by Vibrio fischeri.Mol Microbiol. 2011 Oct;82(1):114-30. doi: 10.1111/j.1365-2958.2011.07800.x. Epub 2011 Aug 30. Mol Microbiol. 2011. PMID: 21854462 Free PMC article.
-
Computational and cellular exploration of the protein-protein interaction between Vibrio fischeri STAS domain protein SypA and serine kinase SypE.Commun Integr Biol. 2023 Apr 20;16(1):2203626. doi: 10.1080/19420889.2023.2203626. eCollection 2023. Commun Integr Biol. 2023. PMID: 37091830 Free PMC article.
-
Control of biofilm formation and colonization in Vibrio fischeri: a role for partner switching?Environ Microbiol. 2010 Aug;12(8):2051-9. doi: 10.1111/j.1462-2920.2010.02269.x. Epub 2010 Jun 9. Environ Microbiol. 2010. PMID: 21966901 Free PMC article. Review.
-
An intricate network of regulators controls biofilm formation and colonization by Vibrio fischeri.Mol Microbiol. 2009 Nov;74(4):782-9. doi: 10.1111/j.1365-2958.2009.06899.x. Epub 2009 Oct 8. Mol Microbiol. 2009. PMID: 19818022 Free PMC article. Review.
Cited by
-
Biofilms 2012: new discoveries and significant wrinkles in a dynamic field.J Bacteriol. 2013 Jul;195(13):2947-58. doi: 10.1128/JB.00239-13. Epub 2013 Apr 26. J Bacteriol. 2013. PMID: 23625847 Free PMC article. Review.
-
The General Stress Response σS Is Regulated by a Partner Switch in the Gram-negative Bacterium Shewanella oneidensis.J Biol Chem. 2016 Dec 9;291(50):26151-26163. doi: 10.1074/jbc.M116.751933. Epub 2016 Nov 3. J Biol Chem. 2016. PMID: 27810894 Free PMC article.
-
Vibrio fischeri Biofilm Formation Prevented by a Trio of Regulators.Appl Environ Microbiol. 2018 Sep 17;84(19):e01257-18. doi: 10.1128/AEM.01257-18. Print 2018 Oct 1. Appl Environ Microbiol. 2018. PMID: 30030225 Free PMC article.
-
Discovery of Calcium as a Biofilm-Promoting Signal for Vibrio fischeri Reveals New Phenotypes and Underlying Regulatory Complexity.J Bacteriol. 2018 Jul 10;200(15):e00016-18. doi: 10.1128/JB.00016-18. Print 2018 Aug 1. J Bacteriol. 2018. PMID: 29463601 Free PMC article.
-
The Histidine Kinase BinK Is a Negative Regulator of Biofilm Formation and Squid Colonization.J Bacteriol. 2016 Sep 9;198(19):2596-607. doi: 10.1128/JB.00037-16. Print 2016 Oct 1. J Bacteriol. 2016. PMID: 26977108 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

