Transcriptome analysis reveals unique metabolic features in the Cryptosporidium parvum Oocysts associated with environmental survival and stresses
- PMID: 23171372
- PMCID: PMC3542205
- DOI: 10.1186/1471-2164-13-647
Transcriptome analysis reveals unique metabolic features in the Cryptosporidium parvum Oocysts associated with environmental survival and stresses
Abstract
Background: Cryptosporidium parvum is a globally distributed zoonotic parasite and an important opportunistic pathogen in immunocompromised patients. Little is known on the metabolic dynamics of the parasite, and study is hampered by the lack of molecular and genetic tools. Here we report the development of the first Agilent microarray for C. parvum (CpArray15K) that covers all predicted ORFs in the parasite genome. Global transcriptome analysis using CpArray15K coupled with real-time qRT-PCR uncovered a number of unique metabolic features in oocysts, the infectious and environmental stage of the parasite.
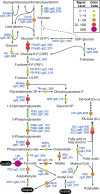
Results: Oocyst stage parasites were found to be highly active in protein synthesis, based on the high transcript levels of genes associated with ribosome biogenesis, transcription and translation. The proteasome and ubiquitin associated components were also highly active, implying that oocysts might employ protein degradation pathways to recycle amino acids in order to overcome the inability to synthesize amino acids de novo. Energy metabolism in oocysts was featured by the highest level of expression of lactate dehydrogenase (LDH) gene. We also studied parasite responses to UV-irradiation, and observed complex and dynamic regulations of gene expression. Notable changes included increased transcript levels of genes involved in DNA repair and intracellular trafficking. Among the stress-related genes, TCP-1 family members and some thioredoxin-associated genes appear to play more important roles in the recovery of UV-induced damages in the oocysts. Our observations also suggest that UV irradiation of oocysts results in increased activities in cytoskeletal rearrangement and intracellular membrane trafficking.
Conclusions: CpArray15K is the first microarray chip developed for C. parvum, which provides the Cryptosporidium research community a needed tool to study the parasite transcriptome and functional genomics. CpArray15K has been successfully used in profiling the gene expressions in the parasite oocysts as well as their responses to UV-irradiation. These observations shed light on how the parasite oocysts might adapt and respond to the hostile external environment and associated stress such as UV irradiation.
Figures


Similar articles
-
Putative cis-regulatory elements associated with heat shock genes activated during excystation of Cryptosporidium parvum.PLoS One. 2010 Mar 4;5(3):e9512. doi: 10.1371/journal.pone.0009512. PLoS One. 2010. PMID: 20209102 Free PMC article.
-
Morpholino-mediated in vivo silencing of Cryptosporidium parvum lactate dehydrogenase decreases oocyst shedding and infectivity.Int J Parasitol. 2018 Jul;48(8):649-656. doi: 10.1016/j.ijpara.2018.01.005. Epub 2018 Mar 9. Int J Parasitol. 2018. PMID: 29530646 Free PMC article.
-
Detection of Cryptosporidium parvum Oocysts on Fresh Produce Using DNA Aptamers.PLoS One. 2015 Sep 3;10(9):e0137455. doi: 10.1371/journal.pone.0137455. eCollection 2015. PLoS One. 2015. PMID: 26334529 Free PMC article.
-
Biology, persistence and detection of Cryptosporidium parvum and Cryptosporidium hominis oocyst.Water Res. 2004 Feb;38(4):818-62. doi: 10.1016/j.watres.2003.10.012. Water Res. 2004. PMID: 14769405 Review.
-
Cryptosporidium parvum and Cyclospora cayetanensis: a review of laboratory methods for detection of these waterborne parasites.J Microbiol Methods. 2002 May;49(3):209-24. doi: 10.1016/s0167-7012(02)00007-6. J Microbiol Methods. 2002. PMID: 11869786 Review.
Cited by
-
Annotated draft genome sequences of three species of Cryptosporidium: Cryptosporidium meleagridis isolate UKMEL1, C. baileyi isolate TAMU-09Q1 and C. hominis isolates TU502_2012 and UKH1.Pathog Dis. 2016 Oct;74(7):ftw080. doi: 10.1093/femspd/ftw080. Epub 2016 Aug 12. Pathog Dis. 2016. PMID: 27519257 Free PMC article.
-
Microbial proteasomes as drug targets.PLoS Pathog. 2021 Dec 9;17(12):e1010058. doi: 10.1371/journal.ppat.1010058. eCollection 2021 Dec. PLoS Pathog. 2021. PMID: 34882737 Free PMC article. Review.
-
Comparative Proteomics Analysis for Elucidating the Interaction Between Host Cells and Toxoplasma gondii.Front Cell Infect Microbiol. 2021 May 13;11:643001. doi: 10.3389/fcimb.2021.643001. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34055664 Free PMC article.
-
Characterization of a Species-Specific Insulinase-Like Protease in Cryptosporidium parvum.Front Microbiol. 2019 Mar 6;10:354. doi: 10.3389/fmicb.2019.00354. eCollection 2019. Front Microbiol. 2019. PMID: 30894838 Free PMC article.
-
Next-Generation Technologies and Systems Biology for the Design of Novel Vaccines Against Apicomplexan Parasites.Front Vet Sci. 2022 Jan 7;8:800361. doi: 10.3389/fvets.2021.800361. eCollection 2021. Front Vet Sci. 2022. PMID: 35071390 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

