Concerted suppression of all starch branching enzyme genes in barley produces amylose-only starch granules
- PMID: 23171412
- PMCID: PMC3537698
- DOI: 10.1186/1471-2229-12-223
Concerted suppression of all starch branching enzyme genes in barley produces amylose-only starch granules
Abstract
Background: Starch is stored in higher plants as granules composed of semi-crystalline amylopectin and amorphous amylose. Starch granules provide energy for the plant during dark periods and for germination of seeds and tubers. Dietary starch is also a highly glycemic carbohydrate being degraded to glucose and rapidly absorbed in the small intestine. But a portion of dietary starch, termed "resistant starch" (RS) escapes digestion and reaches the large intestine, where it is fermented by colonic bacteria producing short chain fatty acids (SCFA) which are linked to several health benefits. The RS is preferentially derived from amylose, which can be increased by suppressing amylopectin synthesis by silencing of starch branching enzymes (SBEs). However all the previous works attempting the production of high RS crops resulted in only partly increased amylose-content and/or significant yield loss.
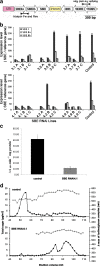
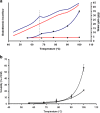
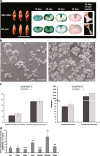
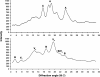
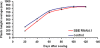
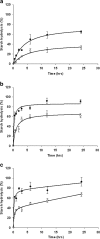
Results: In this study we invented a new method for silencing of multiple genes. Using a chimeric RNAi hairpin we simultaneously suppressed all genes coding for starch branching enzymes (SBE I, SBE IIa, SBE IIb) in barley (Hordeum vulgare L.), resulting in production of amylose-only starch granules in the endosperm. This trait was segregating 3:1. Amylose-only starch granules were irregularly shaped and showed peculiar thermal properties and crystallinity. Transgenic lines retained high-yield possibly due to a pleiotropic upregualtion of other starch biosynthetic genes compensating the SBEs loss. For gelatinized starch, a very high content of RS (65 %) was observed, which is 2.2-fold higher than control (29%). The amylose-only grains germinated with same frequency as control grains. However, initial growth was delayed in young plants.
Conclusions: This is the first time that pure amylose has been generated with high yield in a living organism. This was achieved by a new method of simultaneous suppression of the entire complement of genes encoding starch branching enzymes. We demonstrate that amylopectin is not essential for starch granule crystallinity and integrity. However the slower initial growth of shoots from amylose-only grains may be due to an important physiological role played by amylopectin ordered crystallinity for rapid starch remobilization explaining the broad conservation in the plant kingdom of the amylopectin structure.
Figures
Similar articles
-
Suppressed expression of starch branching enzyme 1 and 2 increases resistant starch and amylose content and modifies amylopectin structure in cassava.Plant Mol Biol. 2022 Mar;108(4-5):413-427. doi: 10.1007/s11103-021-01209-w. Epub 2021 Nov 12. Plant Mol Biol. 2022. PMID: 34767147
-
Control of starch branching in barley defined through differential RNAi suppression of starch branching enzyme IIa and IIb.J Exp Bot. 2010 Mar;61(5):1469-82. doi: 10.1093/jxb/erq011. Epub 2010 Feb 15. J Exp Bot. 2010. PMID: 20156842 Free PMC article.
-
The different effects of starch synthase IIa mutations or variation on endosperm amylose content of barley, wheat and rice are determined by the distribution of starch synthase I and starch branching enzyme IIb between the starch granule and amyloplast stroma.Theor Appl Genet. 2015 Jul;128(7):1407-19. doi: 10.1007/s00122-015-2515-z. Epub 2015 Apr 19. Theor Appl Genet. 2015. PMID: 25893467
-
A review of starch-branching enzymes and their role in amylopectin biosynthesis.IUBMB Life. 2014 Aug;66(8):546-58. doi: 10.1002/iub.1297. Epub 2014 Sep 5. IUBMB Life. 2014. PMID: 25196474 Review.
-
The biosynthesis of starch granules.Biomacromolecules. 2001 Summer;2(2):335-41. doi: 10.1021/bm000133c. Biomacromolecules. 2001. PMID: 11749190 Review.
Cited by
-
High throughput screening of starch structures using carbohydrate microarrays.Sci Rep. 2016 Jul 29;6:30551. doi: 10.1038/srep30551. Sci Rep. 2016. PMID: 27468930 Free PMC article.
-
Suppressed expression of starch branching enzyme 1 and 2 increases resistant starch and amylose content and modifies amylopectin structure in cassava.Plant Mol Biol. 2022 Mar;108(4-5):413-427. doi: 10.1007/s11103-021-01209-w. Epub 2021 Nov 12. Plant Mol Biol. 2022. PMID: 34767147
-
The cereal starch endosperm development and its relationship with other endosperm tissues and embryo.Protoplasma. 2015 Jan;252(1):33-40. doi: 10.1007/s00709-014-0687-z. Epub 2014 Aug 16. Protoplasma. 2015. PMID: 25123370 Review.
-
Starch Granule Re-Structuring by Starch Branching Enzyme and Glucan Water Dikinase Modulation Affects Caryopsis Physiology and Metabolism.PLoS One. 2016 Feb 18;11(2):e0149613. doi: 10.1371/journal.pone.0149613. eCollection 2016. PLoS One. 2016. PMID: 26891365 Free PMC article.
-
Molecular Genetic Analysis of Glucan Branching Enzymes from Plants and Bacteria in Arabidopsis Reveals Marked Differences in Their Functions and Capacity to Mediate Starch Granule Formation.Plant Physiol. 2015 Nov;169(3):1638-55. doi: 10.1104/pp.15.00792. Epub 2015 Sep 10. Plant Physiol. 2015. PMID: 26358415 Free PMC article.
References
-
- BeMiller JN, Whistler RL. Starch: chemistry and technology. USA: Academic Press; 2009.
-
- Badenhuizen N. The biogenesis of starch granules in higher plants. New York: Appleton; 1969.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

