Melanoma antigen-A11 (MAGE-A11) enhances transcriptional activity by linking androgen receptor dimers
- PMID: 23172223
- PMCID: PMC3548502
- DOI: 10.1074/jbc.M112.428409
Melanoma antigen-A11 (MAGE-A11) enhances transcriptional activity by linking androgen receptor dimers
Abstract
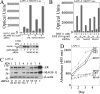
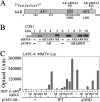
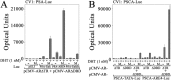
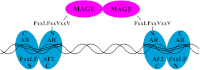
Prostate cancer growth and progression depend on androgen receptor (AR) signaling through transcriptional mechanisms that require interactions with coregulatory proteins, one of which is the primate-specific steroid receptor coregulator melanoma antigen-A11 (MAGE-A11). In this report, we provide evidence how increased expression of MAGE-A11 during prostate cancer progression enhances AR signaling and prostate cancer growth. MAGE-A11 protein levels were highest in castration-recurrent prostate cancer. The cyclic AMP-induced increase in androgen-dependent and androgen-independent AR transcriptional activity correlated with an increase in MAGE-A11 and was inhibited by silencing MAGE-A11 expression. MAGE-A11 mediated synergistic AR transcriptional activity in LAPC-4 prostate cancer cells. The ability of MAGE-A11 to rescue transcriptional activity of complementary inactive AR mutants and promote coimmunoprecipitation between unlike forms of AR suggests that MAGE-A11 links transcriptionally active AR dimers. A model for the AR·MAGE-A11 multidimeric complex is proposed in which one AR FXXLF motif of the AR dimer engages in the androgen-dependent AR NH(2)- and carboxyl-terminal interaction, whereas the second FXXLF motif region of the AR dimer interacts with dimeric MAGE-A11. The AR·MAGE-A11 multidimeric complex accounts for the dual functions of the AR FXXLF motif in the androgen-dependent AR NH(2)- and carboxyl-terminal interaction and binding MAGE-A11 and for synergy between reported AR splice variants and full-length AR. We conclude that the increased expression of MAGE-A11 in castration-recurrent prostate cancer, which is enhanced by cyclic AMP signaling, increases AR-dependent growth of prostate cancer by MAGE-A11 forming a molecular bridge between transcriptionally active AR dimers.
Figures








Similar articles
-
Gain in transcriptional activity by primate-specific coevolution of melanoma antigen-A11 and its interaction site in androgen receptor.J Biol Chem. 2011 Aug 26;286(34):29951-63. doi: 10.1074/jbc.M111.244715. Epub 2011 Jul 5. J Biol Chem. 2011. PMID: 21730049 Free PMC article.
-
Proto-oncogene activity of melanoma antigen-A11 (MAGE-A11) regulates retinoblastoma-related p107 and E2F1 proteins.J Biol Chem. 2013 Aug 23;288(34):24809-24. doi: 10.1074/jbc.M113.468579. Epub 2013 Jul 12. J Biol Chem. 2013. PMID: 23853093 Free PMC article.
-
Up-Regulation of Follistatin-Like 1 By the Androgen Receptor and Melanoma Antigen-A11 in Prostate Cancer.Prostate. 2017 Apr;77(5):505-516. doi: 10.1002/pros.23288. Epub 2016 Dec 14. Prostate. 2017. PMID: 27976415
-
Structural features discriminate androgen receptor N/C terminal and coactivator interactions.Mol Cell Endocrinol. 2012 Jan 30;348(2):403-10. doi: 10.1016/j.mce.2011.03.026. Epub 2011 Jun 1. Mol Cell Endocrinol. 2012. PMID: 21664945 Free PMC article. Review.
-
Functional analysis of androgen receptor N-terminal and ligand binding domain interacting coregulators in prostate cancer.J Formos Med Assoc. 2000 Dec;99(12):885-94. J Formos Med Assoc. 2000. PMID: 11155740 Review.
Cited by
-
Cytotoxic T-lymphocyte elicited therapeutic vaccine candidate targeting cancer against MAGE-A11 carcinogenic protein.Biosci Rep. 2020 Dec 23;40(12):BSR20202349. doi: 10.1042/BSR20202349. Biosci Rep. 2020. PMID: 33169789 Free PMC article.
-
DNA methylation and nucleosome occupancy regulate the cancer germline antigen gene MAGEA11.Epigenetics. 2013 Aug;8(8):849-63. doi: 10.4161/epi.25500. Epub 2013 Jul 9. Epigenetics. 2013. PMID: 23839233 Free PMC article.
-
The link between androgen receptor splice variants and castration-resistant prostate cancer.Horm Cancer. 2014 Aug;5(4):207-17. doi: 10.1007/s12672-014-0177-y. Epub 2014 May 6. Horm Cancer. 2014. PMID: 24798453 Free PMC article. Review.
-
MAGE-A11 expression contributes to cisplatin resistance in head and neck cancer.Clin Oral Investig. 2018 Apr;22(3):1477-1486. doi: 10.1007/s00784-017-2242-8. Epub 2017 Oct 15. Clin Oral Investig. 2018. PMID: 29034444
-
Methylation Profile of X-Chromosome-Related Genes in Male Breast Cancer.Front Oncol. 2020 Jun 17;10:784. doi: 10.3389/fonc.2020.00784. eCollection 2020. Front Oncol. 2020. PMID: 32626651 Free PMC article.
References
-
- Langley E., Zhou Z. X., Wilson E. M. (1995) Evidence for an antiparallel orientation of the ligand activated human androgen receptor dimer. J. Biol. Chem. 270, 29983–29990 - PubMed
-
- He B., Kemppainen J. A., Wilson E. M. (2000) FXXLF and WXXLF sequences mediate the NH2-terminal interaction with the ligand binding domain of the androgen receptor. J. Biol. Chem. 275, 22986–22994 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

