Reconciliation of metabolites and biochemical reactions for metabolic networks
- PMID: 23172809
- PMCID: PMC3896926
- DOI: 10.1093/bib/bbs058
Reconciliation of metabolites and biochemical reactions for metabolic networks
Abstract
Genome-scale metabolic network reconstructions are now routinely used in the study of metabolic pathways, their evolution and design. The development of such reconstructions involves the integration of information on reactions and metabolites from the scientific literature as well as public databases and existing genome-scale metabolic models. The reconciliation of discrepancies between data from these sources generally requires significant manual curation, which constitutes a major obstacle in efforts to develop and apply genome-scale metabolic network reconstructions. In this work, we discuss some of the major difficulties encountered in the mapping and reconciliation of metabolic resources and review three recent initiatives that aim to accelerate this process, namely BKM-react, MetRxn and MNXref (presented in this article). Each of these resources provides a pre-compiled reconciliation of many of the most commonly used metabolic resources. By reducing the time required for manual curation of metabolite and reaction discrepancies, these resources aim to accelerate the development and application of high-quality genome-scale metabolic network reconstructions and models.
Keywords: cheminformatics; data integration; data interoperability; metabolic networks; metabolic resources.
Figures

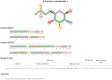

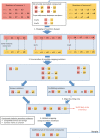

References
-
- Reed JL, Famili I, Thiele I, et al. Towards multidimensional genome annotation. Nat Rev Genet. 2006;7:130–41. - PubMed
-
- Baart GJE, Martens DE. Genome-scale metabolic models: reconstruction and analysis. Methods Mol Biol. 2012;799:107–26. - PubMed
-
- Gianchandani EP, Chavali AK, Papin JA. The application of flux balance analysis in systems biology. Wiley Interdiscip Rev Syst Biol Med. 2010;2:372–82. - PubMed
-
- Stelling J, Klamt S, Bettenbrock K, et al. Metabolic network structure determines key aspects of functionality and regulation. Nature. 2002;420:190–3. - PubMed

