The Saccharomyces cerevisiae F-box protein Dia2 is a mediator of S-phase checkpoint recovery from DNA damage
- PMID: 23172854
- PMCID: PMC3567738
- DOI: 10.1534/genetics.112.146373
The Saccharomyces cerevisiae F-box protein Dia2 is a mediator of S-phase checkpoint recovery from DNA damage
Abstract
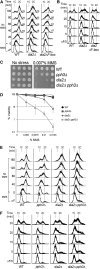
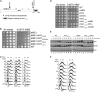
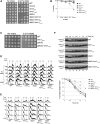
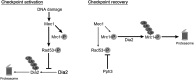
Cell-cycle progression is monitored by checkpoint pathways that pause the cell cycle when stress arises to threaten the integrity of the genome. Although activation of checkpoint pathways has been extensively studied, our understanding of how cells resume the cell cycle when the stress is resolved is relatively limited. In this study, we identify the Saccharomyces cerevisiae F-box protein Dia2 as a novel player in the S-phase checkpoint recovery pathway. Dia2 is required for robust deactivation of the Rad53 checkpoint kinase and timely completion of DNA replication during recovery from DNA damage induced by methyl methanesulfonate (MMS). Aiming to identify the substrate of SCF(Dia2) (Skp1/Cul1/F-box Dia2) in checkpoint recovery, we performed a genetic screen to identify suppressors of dia2Δ cells. The screen identified a new checkpoint-defective allele of MRC1 truncated at the C terminus. We found that checkpoint-defective mrc1 alleles suppress the MMS sensitivity and the checkpoint recovery defect of dia2Δ cells. In addition, Dia2 contributes to Mrc1 degradation during S-phase checkpoint recovery. Furthermore, induced degradation of checkpoint-functional Mrc1 partially rescues the checkpoint recovery defect of dia2Δ cells. We propose a model in which Dia2 mediates Mrc1 degradation to help cells resume the cell cycle during recovery from MMS-induced DNA damage in S-phase.
Figures



Similar articles
-
The F-box protein Dia2 overcomes replication impedance to promote genome stability in Saccharomyces cerevisiae.Genetics. 2006 Dec;174(4):1709-27. doi: 10.1534/genetics.106.057836. Epub 2006 Jun 4. Genetics. 2006. PMID: 16751663 Free PMC article.
-
Activation of the S-phase checkpoint inhibits degradation of the F-box protein Dia2.Mol Cell Biol. 2010 Jan;30(1):160-71. doi: 10.1128/MCB.00612-09. Mol Cell Biol. 2010. PMID: 19858292 Free PMC article.
-
The F-box protein Dia2 regulates DNA replication.Mol Biol Cell. 2006 Apr;17(4):1540-8. doi: 10.1091/mbc.e05-09-0884. Epub 2006 Jan 18. Mol Biol Cell. 2006. PMID: 16421250 Free PMC article.
-
Signaling pathways of replication stress in yeast.FEMS Yeast Res. 2017 Mar 1;17(2). doi: 10.1093/femsyr/fow101. FEMS Yeast Res. 2017. PMID: 27915243 Review.
-
DDR Inc., one business, two associates.Curr Genet. 2019 Apr;65(2):445-451. doi: 10.1007/s00294-018-0908-7. Epub 2018 Nov 22. Curr Genet. 2019. PMID: 30467717 Review.
Cited by
-
Recovery from the DNA Replication Checkpoint.Genes (Basel). 2016 Oct 28;7(11):94. doi: 10.3390/genes7110094. Genes (Basel). 2016. PMID: 27801838 Free PMC article. Review.
-
Proteasome-dependent degradation of replisome components regulates faithful DNA replication.Cell Cycle. 2013 Aug 15;12(16):2564-9. doi: 10.4161/cc.25692. Epub 2013 Jul 18. Cell Cycle. 2013. PMID: 23907116 Free PMC article. Review.
-
The mRNA export adaptor Yra1 contributes to DNA double-strand break repair through its C-box domain.PLoS One. 2019 Apr 5;14(4):e0206336. doi: 10.1371/journal.pone.0206336. eCollection 2019. PLoS One. 2019. PMID: 30951522 Free PMC article.
-
The subunits of the S-phase checkpoint complex Mrc1/Tof1/Csm3: dynamics and interdependence.Cell Div. 2014 Oct 31;9:4. doi: 10.1186/1747-1028-9-4. eCollection 2014. Cell Div. 2014. PMID: 25379053 Free PMC article.
-
Cmr1/WDR76 defines a nuclear genotoxic stress body linking genome integrity and protein quality control.Nat Commun. 2015 Mar 30;6:6533. doi: 10.1038/ncomms7533. Nat Commun. 2015. PMID: 25817432 Free PMC article.
References
-
- Alcasabas A. A., Osborn A. J., Bachant J., Hu F., Werler P. J., et al. , 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3: 958–965 - PubMed
-
- Ang X. L., Harper J.W., 2005. SCF-mediated protein degradation and cell cycle control. Oncogene 24: 2860–2870 - PubMed
-
- Bai C., Sen P., Hofmann K., Ma L., Goebl M., et al. , 1996. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86: 263–274 - PubMed
-
- Bartek J., Lukas J., 2007. DNA damage checkpoints: from initiation to recovery or adaptation. Curr. Opin. Cell Biol. 19: 238–245 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

