New alternately colored FRET sensors for simultaneous monitoring of Zn²⁺ in multiple cellular locations
- PMID: 23173058
- PMCID: PMC3500285
- DOI: 10.1371/journal.pone.0049371
New alternately colored FRET sensors for simultaneous monitoring of Zn²⁺ in multiple cellular locations
Abstract
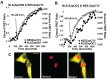
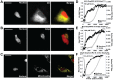
Genetically encoded sensors based on fluorescence resonance energy transfer (FRET) are powerful tools for reporting on ions, molecules and biochemical reactions in living cells. Here we describe the development of new sensors for Zn²⁺based on alternate FRET-pairs that do not involve the traditional CFP and YFP. Zn²⁺ is an essential micronutrient and plays fundamental roles in cell biology. Consequently there is a pressing need for robust sensors to monitor Zn²⁺ levels and dynamics in cells with high spatial and temporal resolution. Here we develop a suite of sensors using alternate FRET pairs, including tSapphire/TagRFP, tSapphire/mKO, Clover/mRuby2, mOrange2/mCherry, and mOrange2/mKATE. These sensors were targeted to both the nucleus and cytosol and characterized and validated in living cells. Sensors based on the new FRET pair Clover/mRuby2 displayed a higher dynamic range and better signal-to-noise ratio than the remaining sensors tested and were optimal for monitoring changes in cytosolic and nuclear Zn²⁺. Using a green-red sensor targeted to the nucleus and cyan-yellow sensor targeted to either the ER, Golgi, or mitochondria, we were able to monitor Zn²⁺ uptake simultaneously in two compartments, revealing that nuclear Zn²⁺ rises quickly, whereas the ER, Golgi, and mitochondria all sequester Zn²⁺ more slowly and with a delay of 600-700 sec. Lastly, these studies provide the first glimpse of nuclear Zn²⁺ and reveal that nuclear Zn²⁺ is buffered at a higher level than cytosolic Zn²⁺.
Conflict of interest statement
Figures



References
-
- Shaner NC, Steinbach PA, Tsien RY (2005) A guide to choosing fluorescent proteins. Nat Methods 2: 905–909. - PubMed
-
- VanEngelenburg SB, Palmer AE (2008) Fluorescent biosensors of protein function. Curr Opin Chem Biol 12: 60–65. - PubMed
-
- Carlson HJ, Campbell RE (2009) Genetically encoded FRET-based biosensors for multiparameter fluorescence imaging. Curr Opin Biotechnol 20: 19–27. - PubMed
-
- Ai HW, Hazelwood KL, Davidson MW, Campbell RE (2008) Fluorescent protein FRET pairs for ratiometric imaging of dual biosensors. Nat Methods 5: 401–403. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

