Familial Alzheimer's disease mutations differentially alter amyloid β-protein oligomerization
- PMID: 23173071
- PMCID: PMC3503339
- DOI: 10.1021/cn300050d
Familial Alzheimer's disease mutations differentially alter amyloid β-protein oligomerization
Abstract
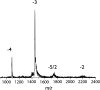
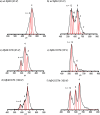
Although most cases of Alzheimer's disease (AD) are sporadic, ∼5% of cases are genetic in origin. These cases, known as familial Alzheimer's disease (FAD), are caused by mutations that alter the rate of production or the primary structure of the amyloid β-protein (Aβ). Changes in the primary structure of Aβ alter the peptide's assembly and toxic activity. Recently, a primary working hypothesis for AD has evolved where causation has been attributed to early, soluble peptide oligomer states. Here we posit that both experimental and pathological differences between FAD-related mutants and wild-type Aβ could be reflected in the early oligomer distributions of these peptides. We use ion mobility-based mass spectrometry to probe the structure and early aggregation states of three mutant forms of Aβ40 and Aβ42: Tottori (D7N), Flemish (A21G), and Arctic (E22G). Our results indicate that the FAD-related amino acid substitutions have no noticeable effect on Aβ monomer cross section, indicating there are no major structural changes in the monomers. However, we observe significant changes to the aggregation states populated by the various Aβ mutants, indicating that structural changes present in the monomers are reflected in the oligomers. Moreover, the early oligomer distributions differ for each mutant, suggesting a possible structural basis for the varied pathogenesis of different forms of FAD.
Figures




References
-
- Dahlgren K. N.; Manelli A. M.; Stine W. B.; Baker L. K.; Krafft G. A.; LaDu M. J. (2002) Oligomeric and fibrillar species of amyloid-β peptides differentially affect neuronal viability. J. Biol. Chem. 277(35), 32046–32053. - PubMed
-
- Bernstein S. L.; Dupuis N. F.; Lazo N. D.; Wyttenbach T.; Condron M. M.; Bitan G.; Teplow D. B.; Shea J. E.; Ruotolo B. T.; Robinson C. V.; Bowers M. T. (2009) Amyloid-β protein oligomerization and the importance of tetramers and dodecamers in the aetiology of Alzheimer’s disease. Nat. Chem. 1(4), 326–331. - PMC - PubMed
- Bernstein S. L.; Wyttenbach T.; Baumketner A.; Shea J. E.; Bitan G.; Teplow D. B.; Bowers M. T. (2005) Amyloid β-protein: Monomer structure and early aggregation states of Aβ42 and its Pro(19) alloform. J. Am. Chem. Soc. 127(7), 2075–2084. - PubMed
- Bitan G.; Kirkitadze M. D.; Lomakin A.; Vollers S. S.; Benedek G. B.; Teplow D. B. (2003) Amyloid β-protein (Aβ) assembly: Aβ40 and Aβ42 oligomerize through distinct pathways. Proc. Natl. Acad. Sci. U.S.A. 100(1), 330–335. - PMC - PubMed
-
- Levylahad E.; Wasco W.; Poorkaj P.; Romano D. M.; Oshima J.; Pettingell W. H.; Yu C. E.; Jondro P. D.; Schmidt S. D.; Wang K.; Crowley A. C.; Fu Y. H.; Guenette S. Y.; Galas D.; Nemens E.; Wijsman E. M.; Bird T. D.; Schellenberg G. D.; Tanzi R. E. (1995) Candidate gene for the chormosome-1 familial Alzheimer’s disease locus. Science 269(5226), 973–977. - PubMed

