Identification of novel genes involved in DNA damage response by screening a genome-wide Schizosaccharomyces pombe deletion library
- PMID: 23173672
- PMCID: PMC3536581
- DOI: 10.1186/1471-2164-13-662
Identification of novel genes involved in DNA damage response by screening a genome-wide Schizosaccharomyces pombe deletion library
Abstract
Background: DNA damage response (DDR) plays pivotal roles in maintaining genome integrity and stability. An effective DDR requires the involvement of hundreds of genes that compose a complicated network. Because DDR is highly conserved in evolution, studies in lower eukaryotes can provide valuable information to elucidate the mechanism in higher organisms. Fission yeast (Schizosaccharomyces pombe) has emerged as an excellent model for DDR research in recent years. To identify novel genes involved in DDR, we screened a genome-wide S. pombe haploid deletion library against six different DNA damage reagents. The library covered 90.5% of the nonessential genes of S. pombe.
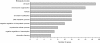
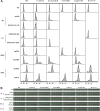
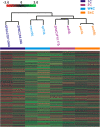
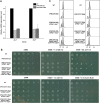
Results: We have identified 52 genes that were actively involved in DDR. Among the 52 genes, 20 genes were linked to DDR for the first time. Flow cytometry analysis of the repair defective mutants revealed that most of them exhibited a defect in cell cycle progression, and some caused genome instability. Microarray analysis and genetic complementation assays were carried out to characterize 6 of the novel DDR genes in more detail. Data suggested that SPBC2A9.02 and SPAC27D7.08c were required for efficient DNA replication initiation because they interacted genetically with DNA replication initiation proteins Abp1 and Abp2. In addition, deletion of sgf73+, meu29+, sec65+ or pab1+ caused improper cytokinesis and DNA re-replication, which contributed to the diploidization in the mutants.
Conclusions: A genome-wide screen of genes involved in DDR emphasized the key role of cell cycle control in the DDR network. Characterization of novel genes identified in the screen helps to elucidate the mechanism of the DDR network and provides valuable clues for understanding genome stability in higher eukaryotes.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

