Competitive amplification of differentially melting amplicons (CADMA) improves KRAS hotspot mutation testing in colorectal cancer
- PMID: 23173730
- PMCID: PMC3517778
- DOI: 10.1186/1471-2407-12-548
Competitive amplification of differentially melting amplicons (CADMA) improves KRAS hotspot mutation testing in colorectal cancer
Abstract
Background: Cancer is an extremely heterogeneous group of diseases traditionally categorized according to tissue of origin. However, even among patients with the same cancer subtype the cellular alterations at the molecular level are often very different. Several new therapies targeting specific molecular changes found in individual patients have initiated the era of personalized therapy and significantly improved patient care. In metastatic colorectal cancer (mCRC) a selected group of patients with wild-type KRAS respond to antibodies against the epidermal growth factor receptor (EGFR). Testing for KRAS mutations is now required prior to anti-EGFR treatment, however, less sensitive methods based on conventional PCR regularly fail to detect KRAS mutations in clinical samples.
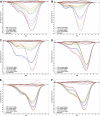
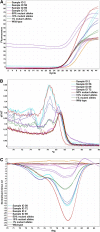
Methods: We have developed sensitive and specific assays for detection of the seven most common KRAS mutations based on a novel methodology named Competitive Amplification of Differentially Melting Amplicons (CADMA). The clinical applicability of these assays was assessed by analyzing 100 colorectal cancer samples, for which KRAS mutation status has been evaluated by the commercially available TheraScreen® KRAS mutation kit.
Results: The CADMA assays were sensitive to at least 0.5% mutant alleles in a wild-type background when using 50 nanograms of DNA in the reactions. Consensus between CADMA and the TheraScreen kit was observed in 96% of the colorectal cancer samples. In cases where disagreement was observed the CADMA result could be confirmed by a previously published assay based on TaqMan probes and by fast COLD-PCR followed by Sanger sequencing.
Conclusions: The high analytical sensitivity and specificity of CADMA may increase diagnostic sensitivity and specificity of KRAS mutation testing in mCRC patients.
Figures
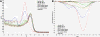
Similar articles
-
Detection of KRAS mutations in colorectal cancer with Fast COLD-PCR.Int J Oncol. 2012 Feb;40(2):378-84. doi: 10.3892/ijo.2011.1221. Epub 2011 Oct 4. Int J Oncol. 2012. PMID: 21971641
-
Low percentage of KRAS mutations revealed by locked nucleic acid polymerase chain reaction: implications for treatment of metastatic colorectal cancer.Mol Med. 2013 Feb 8;18(1):1519-26. doi: 10.2119/molmed.2012.00175. Mol Med. 2013. PMID: 23255073 Free PMC article. Clinical Trial.
-
KRAS analysis in colorectal carcinoma: analytical aspects of Pyrosequencing and allele-specific PCR in clinical practice.BMC Cancer. 2010 Dec 1;10:660. doi: 10.1186/1471-2407-10-660. BMC Cancer. 2010. PMID: 21122130 Free PMC article.
-
KRAS mutation testing in human cancers: The pathologist's role in the era of personalized medicine.Adv Anat Pathol. 2010 Jan;17(1):23-32. doi: 10.1097/PAP.0b013e3181c6962f. Adv Anat Pathol. 2010. PMID: 20032635 Review.
-
Kras in metastatic colorectal cancer.Swiss Med Wkly. 2010 Nov 19;140:w13112. doi: 10.4414/smw.2010.13112. eCollection 2010. Swiss Med Wkly. 2010. PMID: 21104470 Review.
Cited by
-
Colorectal cancer patients with low abundance of KRAS mutation may benefit from EGFR antibody therapy.PLoS One. 2013 Jul 9;8(7):e68022. doi: 10.1371/journal.pone.0068022. Print 2013. PLoS One. 2013. PMID: 23874486 Free PMC article.
-
Mutational Analysis of Circulating Tumor Cells Using a Novel Microfluidic Collection Device and qPCR Assay.Transl Oncol. 2013 Oct 1;6(5):528-38. doi: 10.1593/tlo.13367. eCollection 2013. Transl Oncol. 2013. PMID: 24151533 Free PMC article.
-
Molecular Biomarkers for the Evaluation of Colorectal Cancer.Am J Clin Pathol. 2017 Mar;147(3):221-260. doi: 10.1093/ajcp/aqw209. Epub 2017 Feb 3. Am J Clin Pathol. 2017. PMID: 28165529 Free PMC article.
-
Comprehensive Validation of Snapback Primer-Based Melting Curve Analysis to Detect Nucleotide Variation in the Codon 12 and 13 of KRAS Gene.Biomed Res Int. 2018 Oct 10;2018:8727941. doi: 10.1155/2018/8727941. eCollection 2018. Biomed Res Int. 2018. PMID: 30406144 Free PMC article.
-
Surmounting a PCR challenge using a Contradictory matrix from the Theory of Inventive Problem Solving (TRIZ).Springerplus. 2016 Jan 20;5:56. doi: 10.1186/s40064-015-1577-3. eCollection 2016. Springerplus. 2016. PMID: 26835236 Free PMC article.
References
-
- Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R. et al.Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J clin Oncol: Off J Am Soc Clin Oncol. 2008;26(10):1626–1634. doi: 10.1200/JCO.2007.14.7116. - DOI - PubMed
-
- De Roock W, Jonker DJ, Di Nicolantonio F, Sartore-Bianchi A, Tu D, Siena S, Lamba S, Arena S, Frattini M, Piessevaux H. et al.Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA: J Am Med Assoc. 2010;304(16):1812–1820. doi: 10.1001/jama.2010.1535. - DOI - PubMed
-
- Martini M, Vecchione L, Siena S, Tejpar S, Bardelli A. Targeted therapies: how personal should we go? Nat rev Clin Oncol. 2012;9(2):87–97. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

