RACK1 identified as the PCBP1-interacting protein with a novel functional role on the regulation of human MOR gene expression
- PMID: 23173782
- PMCID: PMC3569494
- DOI: 10.1111/jnc.12100
RACK1 identified as the PCBP1-interacting protein with a novel functional role on the regulation of human MOR gene expression
Abstract
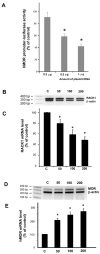
Poly C binding protein 1 (PCBP1) is an expressional regulator of the mu-opioid receptor (MOR) gene. We hypothesized the existence of a PCBP1 co-regulator modifying human MOR gene expression by protein-protein interaction with PCBP1. A human brain cDNA library was screened using the two-hybrid system with PCBP1 as the bait. Receptor for activated protein kinase C (RACK1) protein, containing seven WD domains, was identified. PCBP1-RACK1 interaction was confirmed via in vivo validation using the two-hybrid system, and by co-immunoprecipitation with anti-PCBP1 antibody and human neuronal NMB cell lysate, endogenously expressing PCBP1 and RACK1. Further co-immunoprecipitation suggested that RACK1-PCBP1 interaction occurred in cytosol alone. Single and serial WD domain deletion analyses demonstrated that WD7 of RACK1 is the key domain interacting with PCBP1. RACK1 over-expression resulted in a dose-dependent decrease of MOR promoter activity using p357 plasmid containing human MOR promoter and luciferase reporter gene. Knock-down analysis showed that RACK1 siRNA decreased the endogenous RACK1 mRNA level in NMB, and elevated MOR mRNA level as indicated by RT-PCR. Likewise, a decrease of RACK1 resulted in an increase of MOR proteins, verified by (3) H-diprenorphine binding assay. Collectively, this study reports a novel role of RACK1, physically interacting with PCBP1 and participating in the regulation of human MOR gene expression in neuronal NMB cells.
© 2012 International Society for Neurochemistry.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





 ) and PCBP1 (
) and PCBP1 (
 ) on the left-hand side as well as its product of RACK1-PCBP1 complex on the right-hand side. PCBP1 in the free form enters the nucleus and activates the MOR gene transcription (as depicted in 1). When the equilibrium is shifted to right (more RACK1-PCBP1 complexes are formed) via the RACK1 overexpression, the concentration of free PCBP1 is reduced, so less is available to enter the nucleus, and thus the MOR gene expression is reduced. Conversely, when the availability of RACK1 is reduced by RACK1 siRNA knock down, the equilibrium is shifted to the left (less RACK1-PCBP complex was formed), and thus the concentration of free form of PCBP1 increases, and the MOR gene expression is enhanced at both transcription level (as depicted in 4) and the protein level (as depicted in 5 and 6).
) on the left-hand side as well as its product of RACK1-PCBP1 complex on the right-hand side. PCBP1 in the free form enters the nucleus and activates the MOR gene transcription (as depicted in 1). When the equilibrium is shifted to right (more RACK1-PCBP1 complexes are formed) via the RACK1 overexpression, the concentration of free PCBP1 is reduced, so less is available to enter the nucleus, and thus the MOR gene expression is reduced. Conversely, when the availability of RACK1 is reduced by RACK1 siRNA knock down, the equilibrium is shifted to the left (less RACK1-PCBP complex was formed), and thus the concentration of free form of PCBP1 increases, and the MOR gene expression is enhanced at both transcription level (as depicted in 4) and the protein level (as depicted in 5 and 6).Similar articles
-
Identification of gamma-synuclein as a new PCBP1-interacting protein.Neurol Res. 2016 Dec;38(12):1064-1078. doi: 10.1179/1743132815Y.0000000091. Epub 2016 Oct 27. Neurol Res. 2016. PMID: 26344801
-
Phosphorylation of poly(rC) binding protein 1 (PCBP1) contributes to stabilization of mu opioid receptor (MOR) mRNA via interaction with AU-rich element RNA-binding protein 1 (AUF1) and poly A binding protein (PABP).Gene. 2017 Jan 20;598:113-130. doi: 10.1016/j.gene.2016.11.003. Epub 2016 Nov 9. Gene. 2017. PMID: 27836661 Free PMC article.
-
Post-Transcriptional Regulation of the Human Mu-Opioid Receptor (MOR) by Morphine-Induced RNA Binding Proteins hnRNP K and PCBP1.J Cell Physiol. 2017 Mar;232(3):576-584. doi: 10.1002/jcp.25455. Epub 2016 Jun 24. J Cell Physiol. 2017. PMID: 27292014 Free PMC article.
-
RACK1 vs. HSP90: competition for HIF-1 alpha degradation vs. stabilization.Cell Cycle. 2007 Mar 15;6(6):656-9. doi: 10.4161/cc.6.6.3981. Epub 2007 Mar 7. Cell Cycle. 2007. PMID: 17361105 Review.
-
RACK1, a versatile hub in cancer.Oncogene. 2015 Apr 9;34(15):1890-8. doi: 10.1038/onc.2014.127. Epub 2014 Jun 2. Oncogene. 2015. PMID: 24882575 Review.
Cited by
-
Identification of CD68(+) neutrophil granulocytes in in vitro model of acute inflammation and inflammatory bowel disease.Int J Clin Exp Pathol. 2013;6(4):561-70. Epub 2013 Mar 15. Int J Clin Exp Pathol. 2013. PMID: 23573303 Free PMC article.
-
Transactivation of programmed ribosomal frameshifting by a viral protein.Proc Natl Acad Sci U S A. 2014 May 27;111(21):E2172-81. doi: 10.1073/pnas.1321930111. Epub 2014 May 13. Proc Natl Acad Sci U S A. 2014. PMID: 24825891 Free PMC article.
-
PCBP1 depletion promotes tumorigenesis through attenuation of p27Kip1 mRNA stability and translation.J Exp Clin Cancer Res. 2018 Aug 7;37(1):187. doi: 10.1186/s13046-018-0840-1. J Exp Clin Cancer Res. 2018. PMID: 30086790 Free PMC article.
-
Transcription of the human sodium channel SCN1A gene is repressed by a scaffolding protein RACK1.Mol Neurobiol. 2014 Oct;50(2):438-48. doi: 10.1007/s12035-014-8633-9. Epub 2014 Jan 17. Mol Neurobiol. 2014. PMID: 24436055
-
A novel role for poly(C) binding proteins in programmed ribosomal frameshifting.Nucleic Acids Res. 2016 Jul 8;44(12):5491-503. doi: 10.1093/nar/gkw480. Epub 2016 Jun 2. Nucleic Acids Res. 2016. PMID: 27257056 Free PMC article.
References
-
- Battaini F, Pascale A, Paoletti R, Govoni S. The role of anchoring protein rack1 in pkc activation in the ageing rat brain. Trends in Neurosci. 1997;20:410–415. - PubMed
-
- Baumhaker Y, Ben-Dor T, Bar-Hamburger R, Sarne Y. Characterization of a triple opioid system in the human neuroblastoma NMB cell line. Brain Res. 1994;665:94–100. - PubMed
-
- Buensuceso CS, Woodside D, Huff JL, Plopper GE, O’Toole TE. The WD protein Rack1 mediates protein kinase C and integrin-dependent cell migration. J Cell Sci. 2001;114:1691–1698. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

