TIGRESS: Trustful Inference of Gene REgulation using Stability Selection
- PMID: 23173819
- PMCID: PMC3598250
- DOI: 10.1186/1752-0509-6-145
TIGRESS: Trustful Inference of Gene REgulation using Stability Selection
Abstract
Background: Inferring the structure of gene regulatory networks (GRN) from a collection of gene expression data has many potential applications, from the elucidation of complex biological processes to the identification of potential drug targets. It is however a notoriously difficult problem, for which the many existing methods reach limited accuracy.
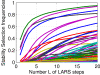
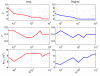
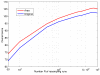
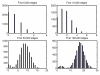
Results: In this paper, we formulate GRN inference as a sparse regression problem and investigate the performance of a popular feature selection method, least angle regression (LARS) combined with stability selection, for that purpose. We introduce a novel, robust and accurate scoring technique for stability selection, which improves the performance of feature selection with LARS. The resulting method, which we call TIGRESS (for Trustful Inference of Gene REgulation with Stability Selection), was ranked among the top GRN inference methods in the DREAM5 gene network inference challenge. In particular, TIGRESS was evaluated to be the best linear regression-based method in the challenge. We investigate in depth the influence of the various parameters of the method, and show that a fine parameter tuning can lead to significant improvements and state-of-the-art performance for GRN inference, in both directed and undirected settings.
Conclusions: TIGRESS reaches state-of-the-art performance on benchmark data, including both in silico and in vivo (E. coli and S. cerevisiae) networks. This study confirms the potential of feature selection techniques for GRN inference. Code and data are available on http://cbio.ensmp.fr/tigress. Moreover, TIGRESS can be run online through the GenePattern platform (GP-DREAM, http://dream.broadinstitute.org).
Figures











Similar articles
-
bLARS: An Algorithm to Infer Gene Regulatory Networks.IEEE/ACM Trans Comput Biol Bioinform. 2016 Mar-Apr;13(2):301-14. doi: 10.1109/TCBB.2015.2450740. IEEE/ACM Trans Comput Biol Bioinform. 2016. PMID: 27045829
-
Gene regulatory network inference using PLS-based methods.BMC Bioinformatics. 2016 Dec 28;17(1):545. doi: 10.1186/s12859-016-1398-6. BMC Bioinformatics. 2016. PMID: 28031031 Free PMC article.
-
Fusing gene expressions and transitive protein-protein interactions for inference of gene regulatory networks.BMC Syst Biol. 2019 Apr 5;13(Suppl 2):37. doi: 10.1186/s12918-019-0695-x. BMC Syst Biol. 2019. PMID: 30953534 Free PMC article.
-
MICRAT: a novel algorithm for inferring gene regulatory networks using time series gene expression data.BMC Syst Biol. 2018 Dec 14;12(Suppl 7):115. doi: 10.1186/s12918-018-0635-1. BMC Syst Biol. 2018. PMID: 30547796 Free PMC article.
-
Biological Network Inference and analysis using SEBINI and CABIN.Methods Mol Biol. 2009;541:551-76. doi: 10.1007/978-1-59745-243-4_24. Methods Mol Biol. 2009. PMID: 19381531 Review.
Cited by
-
Reverse network diffusion to remove indirect noise for better inference of gene regulatory networks.Bioinformatics. 2024 Jul 1;40(7):btae435. doi: 10.1093/bioinformatics/btae435. Bioinformatics. 2024. PMID: 38963312 Free PMC article.
-
Design of Large-Scale Reporter Construct Arrays for Dynamic, Live Cell Systems Biology.ACS Synth Biol. 2018 Sep 21;7(9):2063-2073. doi: 10.1021/acssynbio.8b00236. Epub 2018 Sep 10. ACS Synth Biol. 2018. PMID: 30189139 Free PMC article.
-
Computational Modeling and Reverse Engineering to Reveal Dominant Regulatory Interactions Controlling Osteochondral Differentiation: Potential for Regenerative Medicine.Front Bioeng Biotechnol. 2018 Nov 13;6:165. doi: 10.3389/fbioe.2018.00165. eCollection 2018. Front Bioeng Biotechnol. 2018. PMID: 30483498 Free PMC article. Review.
-
Characterization of DNA Methylation Associated Gene Regulatory Networks During Stomach Cancer Progression.Front Genet. 2019 Feb 4;9:711. doi: 10.3389/fgene.2018.00711. eCollection 2018. Front Genet. 2019. PMID: 30778372 Free PMC article.
-
SABRE: a method for assessing the stability of gene modules in complex tissues and subject populations.BMC Bioinformatics. 2016 Nov 14;17(1):460. doi: 10.1186/s12859-016-1319-8. BMC Bioinformatics. 2016. PMID: 27842512 Free PMC article.
References
-
- Arkin A, Shen P, Ross J. A test case of correlation metric construction of a reaction pathway from measurements. Science. 1997;277(5330):1275–1279. doi: 10.1126/science.277.5330.1275. [ http://www.sciencemag.org/cgi/reprint/277/5330/1275.pdf] - DOI
-
- Liang S, Fuhrman S, Somogyi R. REVEAL, a general reverse engineering algorithm for inference of genetic network architectures. Pac Symp Biocomput. 1998;3:18–29. - PubMed
-
- Chen T, He HL, Church GM. Modeling gene expression with differential equations. Pac Symp Biocomput. 1999;4:29–40. - PubMed
-
- Yeung MKS, Tegnér J, Collins JJ. Reverse engineering gene networks using singular value decomposition and robust regression. Proc Natl Acad Sci USA. 2002;99(9):6163–6168. doi: 10.1073/pnas.092576199. [ http://www.pnas.org/content/99/9/6163.abstract] - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

