Discrete nature of EpCAM+ and CD90+ cancer stem cells in human hepatocellular carcinoma
- PMID: 23174907
- PMCID: PMC7180389
- DOI: 10.1002/hep.26168
Discrete nature of EpCAM+ and CD90+ cancer stem cells in human hepatocellular carcinoma
Abstract
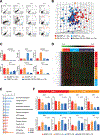
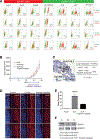
Recent evidence suggests that hepatocellular carcinoma (HCC) is organized by a subset of cells with stem cell features (cancer stem cells; CSCs). CSCs are considered a pivotal target for the eradication of cancer, and liver CSCs have been identified by the use of various stem cell markers. However, little information is known about the expression patterns and characteristics of marker-positive CSCs, hampering the development of personalized CSC-targeted therapy. Here, we show that CSC markers EpCAM and CD90 are independently expressed in liver cancer. In primary HCC, EpCAM+ and CD90+ cells resided distinctively, and gene-expression analysis of sorted cells suggested that EpCAM+ cells had features of epithelial cells, whereas CD90+ cells had those of vascular endothelial cells. Clinicopathological analysis indicated that the presence of EpCAM+ cells was associated with poorly differentiated morphology and high serum alpha-fetoprotein (AFP), whereas the presence of CD90+ cells was associated with a high incidence of distant organ metastasis. Serial xenotransplantation of EpCAM+ /CD90+ cells from primary HCCs in immune-deficient mice revealed rapid growth of EpCAM+ cells in the subcutaneous lesion and a highly metastatic capacity of CD90+ cells in the lung. In cell lines, CD90+ cells showed abundant expression of c-Kit and in vitro chemosensitivity to imatinib mesylate. Furthermore, CD90+ cells enhanced the motility of EpCAM+ cells when cocultured in vitro through the activation of transforming growth factor beta (TGF-β) signaling, whereas imatinib mesylate suppressed TGFB1 expression in CD90+ cells as well as CD90+ cell-induced motility of EpCAM+ cells.
Conclusion: Our data suggest the discrete nature and potential interaction of EpCAM+ and CD90+ CSCs with specific gene-expression patterns and chemosensitivity to molecular targeted therapy. The presence of distinct CSCs may determine the clinical outcome of HCC.
Copyright © 2012 American Association for the Study of Liver Diseases.
Figures




References
-
- Chiba T, Kita K, Zheng YW, Yokosuka O, Saisho H, Iwama A, et al. Side population purified from hepatocellular carcinoma cells harbors cancer stem cell-like properties. HEPATOLOGY 2006;44:240–251. - PubMed
-
- Ma S, Tang KH, Chan YP, Lee TK, Kwan PS, Castilho A, et al. miR130b promotes CD133(+) liver tumor-initiating cell growth and self-renewal via tumor protein 53-induced nuclear protein 1. Cell Stem Cell 2010;7:694–707. - PubMed
-
- Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai P, et al. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell 2008;13:153–166. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
