Subcellular localization and function of an epitope-tagged p7 viroporin in hepatitis C virus-producing cells
- PMID: 23175364
- PMCID: PMC3554161
- DOI: 10.1128/JVI.02782-12
Subcellular localization and function of an epitope-tagged p7 viroporin in hepatitis C virus-producing cells
Abstract
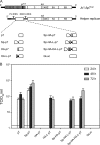
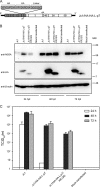
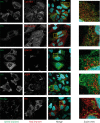
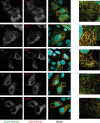
The hepatitis C virus (HCV) viroporin p7 is crucial for production of infectious viral progeny. However, its role in the viral replication cycle remains incompletely understood, in part due to the poor availability of p7-specific antibodies. To circumvent this obstacle, we inserted two consecutive hemagglutinin (HA) epitope tags at its N terminus. HA-tagged p7 reduced peak virus titers ca. 10-fold and decreased kinetics of virus production compared to the wild-type virus. However, HA-tagged p7 rescued virus production of a mutant virus lacking p7, thus providing formal proof that the tag does not disrupt p7 function. In HCV-producing cells, p7 displayed a reticular staining pattern which colocalized with the HCV envelope glycoprotein 2 (E2) but also partially with viral nonstructural proteins 2, 3, and 5A. Using coimmunoprecipitation, we confirmed a specific interaction between p7 and NS2, whereas we did not detect a stable interaction with core, E2, or NS5A. Moreover, we did not observe p7 incorporation into affinity-purified virus particles. Consistently, there was no evidence supporting a role of p7 in viral entry, as an anti-HA antibody was not able to neutralize Jc1 virus produced from an HA-p7-tagged genome. Collectively, these findings highlight a stable interaction between p7 and NS2 which is likely crucial for production of infectious HCV particles. Use of this functional epitope-tagged p7 variant should facilitate the analysis of the final steps of the HCV replication cycle.
Figures
Similar articles
-
Detergent-resistant membrane association of NS2 and E2 during hepatitis C virus replication.J Virol. 2015 Apr;89(8):4562-74. doi: 10.1128/JVI.00123-15. Epub 2015 Feb 11. J Virol. 2015. PMID: 25673706 Free PMC article.
-
NS2 protein of hepatitis C virus interacts with structural and non-structural proteins towards virus assembly.PLoS Pathog. 2011 Feb 10;7(2):e1001278. doi: 10.1371/journal.ppat.1001278. PLoS Pathog. 2011. PMID: 21347350 Free PMC article.
-
The subcellular localization of the hepatitis C virus non-structural protein NS2 is regulated by an ion channel-independent function of the p7 protein.J Gen Virol. 2011 Apr;92(Pt 4):819-30. doi: 10.1099/vir.0.027441-0. Epub 2010 Dec 22. J Gen Virol. 2011. PMID: 21177929 Free PMC article.
-
Structural and Functional Properties of the Hepatitis C Virus p7 Viroporin.Viruses. 2015 Aug 6;7(8):4461-81. doi: 10.3390/v7082826. Viruses. 2015. PMID: 26258788 Free PMC article. Review.
-
Processing and functions of Hepatitis C virus proteins.Intervirology. 1999;42(2-3):145-52. doi: 10.1159/000024973. Intervirology. 1999. PMID: 10516468 Review.
Cited by
-
Three-dimensional structure and interaction studies of hepatitis C virus p7 in 1,2-dihexanoyl-sn-glycero-3-phosphocholine by solution nuclear magnetic resonance.Biochemistry. 2013 Aug 6;52(31):5295-303. doi: 10.1021/bi4006623. Epub 2013 Jul 24. Biochemistry. 2013. PMID: 23841474 Free PMC article.
-
Ion Channel Function and Cross-Species Determinants in Viral Assembly of Nonprimate Hepacivirus p7.J Virol. 2016 Apr 29;90(10):5075-5089. doi: 10.1128/JVI.00132-16. Print 2016 May 15. J Virol. 2016. PMID: 26962224 Free PMC article.
-
ABHD5/CGI-58, the Chanarin-Dorfman Syndrome Protein, Mobilises Lipid Stores for Hepatitis C Virus Production.PLoS Pathog. 2016 Apr 28;12(4):e1005568. doi: 10.1371/journal.ppat.1005568. eCollection 2016 Apr. PLoS Pathog. 2016. PMID: 27124600 Free PMC article.
-
The intraviral protein interaction network of hepatitis C virus.Mol Cell Proteomics. 2014 Jul;13(7):1676-89. doi: 10.1074/mcp.M113.036301. Epub 2014 May 5. Mol Cell Proteomics. 2014. PMID: 24797426 Free PMC article.
-
Hepatitis C virus, mitochondria and auto/mitophagy: exploiting a host defense mechanism.World J Gastroenterol. 2014 Mar 14;20(10):2624-33. doi: 10.3748/wjg.v20.i10.2624. World J Gastroenterol. 2014. PMID: 24627598 Free PMC article. Review.
References
-
- Griffin SD, Beales LP, Clarke DS, Worsfold O, Evans SD, Jaeger J, Harris MP, Rowlands DJ. 2003. The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, amantadine. FEBS Lett. 535:34–38 - PubMed
-
- Premkumar A, Wilson L, Ewart GD, Gage PW. 2004. Cation-selective ion channels formed by p7 of hepatitis C virus are blocked by hexamethylene amiloride. FEBS Lett. 557:99–103 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

