OmpR regulation of the uropathogenic Escherichia coli fimB gene in an acidic/high osmolality environment
- PMID: 23175504
- PMCID: PMC3709560
- DOI: 10.1099/mic.0.059386-0
OmpR regulation of the uropathogenic Escherichia coli fimB gene in an acidic/high osmolality environment
Abstract
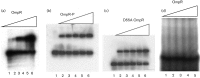
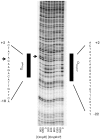
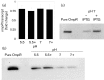
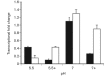
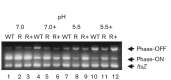
Uropathogenic Escherichia coli (UPEC) causes more than 90 % of all human urinary tract infections through type 1 piliated UPEC cells binding to bladder epithelial cells. The FimB and FimE site-specific recombinases orient the fimS element containing the fimA structural gene promoter. Regulation of fimB and fimE depends on environmental pH and osmolality. The EnvZ/OmpR two-component system affects osmoregulation in E. coli. To ascertain if OmpR directly regulated the fimB gene promoters, gel mobility shift and DNase I footprinting experiments were performed using OmpR or phosphorylated OmpR (OmpR-P) mixed with the fimB promoter regions of UPEC strain NU149. Both OmpR-P and OmpR bound weakly to one fimB promoter. Because there was weak binding to one fimB promoter, strain NU149 was grown in different pH and osmolality environments, and total RNAs were extracted from each population and converted to cDNAs. Quantitative reverse-transcriptase PCR showed no differences in ompR transcription among the different growth conditions. Conversely, Western blots showed a significant increase in OmpR protein in UPEC cells grown in a combined low pH/high osmolality environment versus a neutral pH/high osmolality environment. In a high osmolality environment, the ompR mutant expressed more fimB transcripts and Phase-ON positioning of the fimS element as well as higher type 1 pili levels than wild-type cells. Together these results suggest that OmpR may be post-transcriptionally regulated in UPEC cells growing in a low pH/high osmolality environment, which regulates fimB in UPEC.
Figures
Similar articles
-
Osmolarity and pH growth conditions regulate fim gene transcription and type 1 pilus expression in uropathogenic Escherichia coli.Infect Immun. 2002 Mar;70(3):1391-402. doi: 10.1128/IAI.70.3.1391-1402.2002. Infect Immun. 2002. PMID: 11854225 Free PMC article.
-
Regulation of Escherichia coli fim gene transcription by GadE and other acid tolerance gene products.Microbiology (Reading). 2022 Mar;168(3):001149. doi: 10.1099/mic.0.001149. Microbiology (Reading). 2022. PMID: 35316170 Free PMC article.
-
RfaH suppresses small RNA MicA inhibition of fimB expression in Escherichia coli K-12.J Bacteriol. 2014 Jan;196(1):148-56. doi: 10.1128/JB.00912-13. Epub 2013 Oct 25. J Bacteriol. 2014. PMID: 24163336 Free PMC article.
-
Comprehensive analysis of type 1 fimbriae regulation in fimB-null strains from the multidrug resistant Escherichia coli ST131 clone.Mol Microbiol. 2016 Sep;101(6):1069-87. doi: 10.1111/mmi.13442. Epub 2016 Jul 15. Mol Microbiol. 2016. PMID: 27309594
-
Temporal Regulation of fim Genes in Uropathogenic Escherichia coli during Infection of the Murine Urinary Tract.J Pathog. 2017;2017:8694356. doi: 10.1155/2017/8694356. Epub 2017 Dec 27. J Pathog. 2017. PMID: 29445547 Free PMC article.
Cited by
-
In Vivo Role of Two-Component Regulatory Systems in Models of Urinary Tract Infections.Pathogens. 2023 Jan 10;12(1):119. doi: 10.3390/pathogens12010119. Pathogens. 2023. PMID: 36678467 Free PMC article. Review.
-
CRISPRi-mediated suppression of E. coli Nissle 1917 virulence factors: A strategy for creating an engineered probiotic using csgD gene suppression.Front Nutr. 2022 Aug 1;9:938989. doi: 10.3389/fnut.2022.938989. eCollection 2022. Front Nutr. 2022. PMID: 35978963 Free PMC article.
-
Bacterial resistance to leucyl-tRNA synthetase inhibitor GSK2251052 develops during treatment of complicated urinary tract infections.Antimicrob Agents Chemother. 2015 Jan;59(1):289-98. doi: 10.1128/AAC.03774-14. Epub 2014 Oct 27. Antimicrob Agents Chemother. 2015. PMID: 25348524 Free PMC article. Clinical Trial.
-
sRNA-Mediated Regulation of P-Fimbriae Phase Variation in Uropathogenic Escherichia coli.PLoS Pathog. 2015 Aug 20;11(8):e1005109. doi: 10.1371/journal.ppat.1005109. eCollection 2015 Aug. PLoS Pathog. 2015. PMID: 26291711 Free PMC article.
-
Unexpected properties of sRNA promoters allow feedback control via regulation of a two-component system.Nucleic Acids Res. 2016 Nov 16;44(20):9650-9666. doi: 10.1093/nar/gkw642. Epub 2016 Jul 20. Nucleic Acids Res. 2016. PMID: 27439713 Free PMC article.
References
-
- Abraham S. N., Babu J. P., Giampapa C. S., Hasty D. L., Simpson W. A., Beachey E. H. (1985a). Protection against Escherichia coli-induced urinary tract infections with hybridoma antibodies directed against type 1 fimbriae or complementary d-mannose receptors. Infect Immun 48, 625–628. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

