Structure, phosphorylation and U2AF65 binding of the N-terminal domain of splicing factor 1 during 3'-splice site recognition
- PMID: 23175611
- PMCID: PMC3553976
- DOI: 10.1093/nar/gks1097
Structure, phosphorylation and U2AF65 binding of the N-terminal domain of splicing factor 1 during 3'-splice site recognition
Abstract
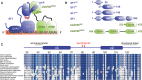
Recognition of the 3'-splice site is a key step in pre-mRNA splicing and accomplished by a dynamic complex comprising splicing factor 1 (SF1) and the U2 snRNP auxiliary factor 65-kDa subunit (U2AF65). Both proteins mediate protein-protein and protein-RNA interactions for cooperative RNA-binding during spliceosome assembly. Here, we report the solution structure of a novel helix-hairpin domain in the N-terminal region of SF1 (SF1(NTD)). The nuclear magnetic resonance- and small-angle X-ray scattering-derived structure of a complex of the SF1(NTD) with the C-terminal U2AF homology motif domain of U2AF65 (U2AF65(UHM)) reveals that, in addition to the known U2AF65(UHM)-SF1 interaction, the helix-hairpin domain forms a secondary, hydrophobic interface with U2AF65(UHM), which locks the orientation of the two subunits. Mutational analysis shows that the helix hairpin is essential for cooperative formation of the ternary SF1-U2AF65-RNA complex. We further show that tandem serine phosphorylation of a conserved Ser80-Pro81-Ser82-Pro83 motif rigidifies a long unstructured linker in the SF1 helix hairpin. Phosphorylation does not significantly alter the overall conformations of SF1, SF1-U2AF65 or the SF1-U2AF65-RNA complexes, but slightly enhances RNA binding. Our results indicate that the helix-hairpin domain of SF1 is required for cooperative 3'-splice site recognition presumably by stabilizing a unique quaternary arrangement of the SF1-U2AF65-RNA complex.
Figures



Similar articles
-
Multiple U2AF65 binding sites within SF3b155: thermodynamic and spectroscopic characterization of protein-protein interactions among pre-mRNA splicing factors.J Mol Biol. 2006 Feb 24;356(3):664-83. doi: 10.1016/j.jmb.2005.11.067. Epub 2005 Dec 7. J Mol Biol. 2006. PMID: 16376933 Free PMC article.
-
Major phosphorylation of SF1 on adjacent Ser-Pro motifs enhances interaction with U2AF65.FEBS J. 2006 Feb;273(3):577-87. doi: 10.1111/j.1742-4658.2005.05091.x. FEBS J. 2006. PMID: 16420481 Free PMC article.
-
Structure of phosphorylated SF1 bound to U2AF⁶⁵ in an essential splicing factor complex.Structure. 2013 Feb 5;21(2):197-208. doi: 10.1016/j.str.2012.10.020. Epub 2012 Dec 27. Structure. 2013. PMID: 23273425 Free PMC article.
-
U2AF homology motifs: protein recognition in the RRM world.Genes Dev. 2004 Jul 1;18(13):1513-26. doi: 10.1101/gad.1206204. Genes Dev. 2004. PMID: 15231733 Free PMC article. Review.
-
The structure of human cleavage factor I(m) hints at functions beyond UGUA-specific RNA binding: a role in alternative polyadenylation and a potential link to 5' capping and splicing.RNA Biol. 2011 Sep-Oct;8(5):748-53. doi: 10.4161/rna.8.5.16040. Epub 2011 Sep 1. RNA Biol. 2011. PMID: 21881408 Free PMC article. Review.
Cited by
-
Quantitative phosphoproteomics analysis of nitric oxide-responsive phosphoproteins in cotton leaf.PLoS One. 2014 Apr 8;9(4):e94261. doi: 10.1371/journal.pone.0094261. eCollection 2014. PLoS One. 2014. PMID: 24714030 Free PMC article.
-
Structure-function analysis of the 5' end of yeast U1 snRNA highlights genetic interactions with the Msl5*Mud2 branchpoint-binding complex and other spliceosome assembly factors.Nucleic Acids Res. 2013 Aug;41(15):7485-500. doi: 10.1093/nar/gkt490. Epub 2013 Jun 10. Nucleic Acids Res. 2013. PMID: 23754852 Free PMC article.
-
RNA-Binding Proteins: Splicing Factors and Disease.Biomolecules. 2015 May 13;5(2):893-909. doi: 10.3390/biom5020893. Biomolecules. 2015. PMID: 25985083 Free PMC article. Review.
-
Modulation of HIV-1 gene expression by binding of a ULM motif in the Rev protein to UHM-containing splicing factors.Nucleic Acids Res. 2019 May 21;47(9):4859-4871. doi: 10.1093/nar/gkz185. Nucleic Acids Res. 2019. PMID: 30892606 Free PMC article.
-
Cancer-Associated Mutations Mapped on High-Resolution Structures of the U2AF2 RNA Recognition Motifs.Biochemistry. 2017 Sep 12;56(36):4757-4761. doi: 10.1021/acs.biochem.7b00551. Epub 2017 Sep 1. Biochemistry. 2017. PMID: 28850223 Free PMC article.
References
-
- Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. - PubMed
-
- Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

