Linking DNA methyltransferases to epigenetic marks and nucleosome structure genome-wide in human tumor cells
- PMID: 23177624
- PMCID: PMC3625945
- DOI: 10.1016/j.celrep.2012.10.017
Linking DNA methyltransferases to epigenetic marks and nucleosome structure genome-wide in human tumor cells
Abstract
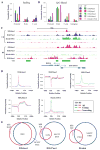
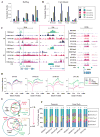
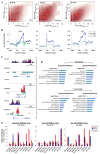
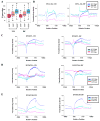
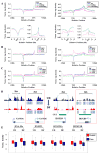
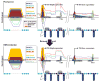
DNA methylation, mediated by the combined action of three DNA methyltransferases (DNMT1, DNMT3A, and DNMT3B), is essential for mammalian development and is a major contributor to cellular transformation. To elucidate how DNA methylation is targeted, we mapped the genome-wide localization of all DNMTs and methylation, and examined the relationships among these markers, histone modifications, and nucleosome structure in a pluripotent human tumor cell line in its undifferentiated and differentiated states. Our findings reveal a strong link between DNMTs and transcribed loci, and that DNA methylation is not a simple sum of DNMT localization patterns. A comparison of the epigenomes of normal and cancerous stem cells, and pluripotent and differentiated states shows that the presence of at least two DNMTs is strongly associated with loci targeted for DNA hypermethylation. Taken together, these results shed important light on the determinants of DNA methylation and how it may become disrupted in cancer cells.
Copyright © 2012 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
DNA methylation determines nucleosome occupancy in the 5'-CpG islands of tumor suppressor genes.Oncogene. 2013 Nov 21;32(47):5421-8. doi: 10.1038/onc.2013.162. Epub 2013 May 20. Oncogene. 2013. PMID: 23686312 Free PMC article.
-
Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation.Nature. 2015 Apr 9;520(7546):243-7. doi: 10.1038/nature14176. Epub 2015 Jan 21. Nature. 2015. PMID: 25607372
-
DNMT1 and DNMT3B modulate distinct polycomb-mediated histone modifications in colon cancer.Cancer Res. 2009 Sep 15;69(18):7412-21. doi: 10.1158/0008-5472.CAN-09-0116. Epub 2009 Sep 1. Cancer Res. 2009. PMID: 19723660 Free PMC article.
-
Genetic Studies on Mammalian DNA Methyltransferases.Adv Exp Med Biol. 2016;945:123-150. doi: 10.1007/978-3-319-43624-1_6. Adv Exp Med Biol. 2016. PMID: 27826837 Review.
-
DNA methyltransferases hitchhiking on chromatin.Swiss Med Wkly. 2020 Aug 24;150:w20329. doi: 10.4414/smw.2020.20329. eCollection 2020 Aug 24. Swiss Med Wkly. 2020. PMID: 32920789 Review.
Cited by
-
Parent-of-origin effects of A1CF and AGO2 on testicular germ-cell tumors, testicular abnormalities, and fertilization bias.Proc Natl Acad Sci U S A. 2016 Sep 13;113(37):E5425-33. doi: 10.1073/pnas.1604773113. Epub 2016 Aug 31. Proc Natl Acad Sci U S A. 2016. PMID: 27582469 Free PMC article.
-
Differential DNA methylation with age displays both common and dynamic features across human tissues that are influenced by CpG landscape.Genome Biol. 2013;14(9):R102. doi: 10.1186/gb-2013-14-9-r102. Genome Biol. 2013. PMID: 24034465 Free PMC article.
-
Active DNA demethylation is upstream of rod-photoreceptor fate determination and required for retinal development.bioRxiv [Preprint]. 2025 Feb 3:2025.02.03.636318. doi: 10.1101/2025.02.03.636318. bioRxiv. 2025. Update in: PLoS Biol. 2025 Aug 4;23(8):e3003332. doi: 10.1371/journal.pbio.3003332. PMID: 39975078 Free PMC article. Updated. Preprint.
-
Genome-wide positioning of bivalent mononucleosomes.BMC Med Genomics. 2016 Sep 15;9(1):60. doi: 10.1186/s12920-016-0221-6. BMC Med Genomics. 2016. PMID: 27634286 Free PMC article.
-
Targeting the epigenome of cancer stem cells in pediatric nervous system tumors.Mol Cell Biochem. 2023 Oct;478(10):2241-2255. doi: 10.1007/s11010-022-04655-2. Epub 2023 Jan 13. Mol Cell Biochem. 2023. PMID: 36637615 Review.
References
-
- Adamo A, Sese B, Boue S, Castano J, Paramonov I, Barrero MJ, Izpisua Belmonte JC. LSD1 regulates the balance between self-renewal and differentiation in human embryonic stem cells. Nat Cell Biol. 2011;13:652–659. - PubMed
-
- Aran D, Toperoff G, Rosenberg M, Hellman A. Replication timing-related and gene body-specific methylation of active human genes. Hum Mol Genet. 2011;20:670–680. - PubMed
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
-
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

