ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility
- PMID: 23177736
- PMCID: PMC3646334
- DOI: 10.1016/j.molcel.2012.10.015
ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility
Abstract
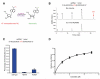
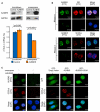
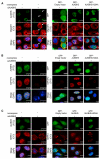
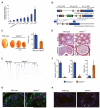
N(6)-methyladenosine (m(6)A) is the most prevalent internal modification of messenger RNA (mRNA) in higher eukaryotes. Here we report ALKBH5 as another mammalian demethylase that oxidatively reverses m(6)A in mRNA in vitro and in vivo. This demethylation activity of ALKBH5 significantly affects mRNA export and RNA metabolism as well as the assembly of mRNA processing factors in nuclear speckles. Alkbh5-deficient male mice have increased m(6)A in mRNA and are characterized by impaired fertility resulting from apoptosis that affects meiotic metaphase-stage spermatocytes. In accordance with this defect, we have identified in mouse testes 1,551 differentially expressed genes that cover broad functional categories and include spermatogenesis-related mRNAs involved in the p53 functional interaction network. The discovery of this RNA demethylase strongly suggests that the reversible m(6)A modification has fundamental and broad functions in mammalian cells.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures

Comment in
-
Sprouts of RNA epigenetics: the discovery of mammalian RNA demethylases.RNA Biol. 2013 Jun;10(6):915-8. doi: 10.4161/rna.24711. Epub 2013 Apr 17. RNA Biol. 2013. PMID: 23619745 Free PMC article.
References
-
- Achanzar WE, Ward S. A nematode gene required for sperm vesicle fusion. J. Cell Sci. 1997;110:1073–1081. - PubMed
-
- Baltz AG, Munschauer M, Schwanhaüsser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell. 2012;46:674–690. - PubMed
-
- Bokar JA. In: Fine-Tuning of RNA Functions by Modification and Editing. Grosjean H, editor. Volume 12. Springer; New York: 2005. pp. 141–177.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

