Genome-wide methylation profiles reveal quantitative views of human aging rates
- PMID: 23177740
- PMCID: PMC3780611
- DOI: 10.1016/j.molcel.2012.10.016
Genome-wide methylation profiles reveal quantitative views of human aging rates
Abstract
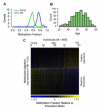
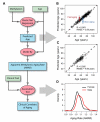
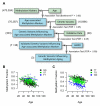
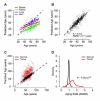
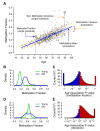
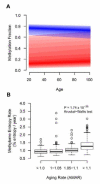
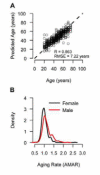
The ability to measure human aging from molecular profiles has practical implications in many fields, including disease prevention and treatment, forensics, and extension of life. Although chronological age has been linked to changes in DNA methylation, the methylome has not yet been used to measure and compare human aging rates. Here, we build a quantitative model of aging using measurements at more than 450,000 CpG markers from the whole blood of 656 human individuals, aged 19 to 101. This model measures the rate at which an individual's methylome ages, which we show is impacted by gender and genetic variants. We also show that differences in aging rates help explain epigenetic drift and are reflected in the transcriptome. Moreover, we show how our aging model is upheld in other human tissues and reveals an advanced aging rate in tumor tissue. Our model highlights specific components of the aging process and provides a quantitative readout for studying the role of methylation in age-related disease.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
Comment in
-
Human epigenetics: Showing your age.Nat Rev Genet. 2013 Jan;14(1):6. doi: 10.1038/nrg3391. Epub 2012 Dec 4. Nat Rev Genet. 2013. PMID: 23207909 No abstract available.
References
-
- Alisch RS, Barwick BG, Chopra P, Myrick LK, Satten GA, Conneely KN, Warren ST. [Accessed May 17, 2012];Age-associated DNA methylation in pediatric populations. Genome Research. 2012 22:623–632. Available at: http://www.ncbi.nlm.nih.gov/pubmed/22300631. - PMC - PubMed
-
- Atzmon G, Rincon M, Schechter CB, Shuldiner AR, Lipton RB, Bergman A, Barzilai N. [Accessed January 18, 2012];Lipoprotein Genotype and Conserved Pathway for Exceptional Longevity in Humans. PLoS Biol. 2006 4:e113. Available at: UR http://dx.doi.org/10.1371/journal.pbio.0040113,http://dx.doi.org/10.1371.... - DOI - PMC - PubMed
-
- Barres R, Zierath JR. [Accessed January 18, 2012];DNA methylation in metabolic disorders. The American Journal of Clinical Nutrition. 2011 Available at: http://www.ajcn.org/content/early/2011/02/02/ajcn.110.001933.abstract. - PubMed
-
- Bell JT, Pai AA, Pickrell JK, Gaffney DJ, Pique-Regi R, Degner JF, Gilad Y, Pritchard JK. [Accessed January 31, 2012];DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biology. 2011 12:R10. Available at: http://www.ncbi.nlm.nih.gov/pubmed/21251332. - PMC - PubMed
-
- Bell JT, Tsai P-C, Yang T-P, Pidsley R, Nisbet J, Glass D, Mangino M, Zhai G, Zhang F, Valdes A, et al. [Accessed May 17, 2012];Epigenome-Wide Scans Identify Differentially Methylated Regions for Age and Age-Related Phenotypes in a Healthy Ageing Population. PLoS Genet. 2012 8:e1002629. Available at: http://dx.doi.org/10.1371/journal.pgen.1002629. - DOI - PMC - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
- P30 MH062261/MH/NIMH NIH HHS/United States
- P50GM085764,/GM/NIGMS NIH HHS/United States
- R01 EY014428/EY/NEI NIH HHS/United States
- R01E5014811/PHS HHS/United States
- P50 GM085764/GM/NIGMS NIH HHS/United States
- EY021374/EY/NEI NIH HHS/United States
- EY018660/EY/NEI NIH HHS/United States
- EY019270/EY/NEI NIH HHS/United States
- EY014428/EY/NEI NIH HHS/United States
- R01 EY021374/EY/NEI NIH HHS/United States
- R01 ES014811/ES/NIEHS NIH HHS/United States
- R01 EY019270/EY/NEI NIH HHS/United States
- T32 GM007198/GM/NIGMS NIH HHS/United States
- R01 EY018660/EY/NEI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
