Endocardial cells form the coronary arteries by angiogenesis through myocardial-endocardial VEGF signaling
- PMID: 23178125
- PMCID: PMC3508471
- DOI: 10.1016/j.cell.2012.10.023
Endocardial cells form the coronary arteries by angiogenesis through myocardial-endocardial VEGF signaling
Abstract
The origins and developmental mechanisms of coronary arteries are incompletely understood. We show here by fate mapping, clonal analysis, and immunohistochemistry that endocardial cells generate the endothelium of coronary arteries. Dye tracking, live imaging, and tissue transplantation also revealed that ventricular endocardial cells are not terminally differentiated; instead, they are angiogenic and form coronary endothelial networks. Myocardial Vegf-a or endocardial Vegfr-2 deletion inhibited coronary angiogenesis and arterial formation by ventricular endocardial cells. In contrast, lineage and knockout studies showed that endocardial cells make a small contribution to the coronary veins, the formation of which is independent of myocardial-to-endocardial Vegf signaling. Thus, contrary to the current view of a common source for the coronary vessels, our findings indicate that the coronary arteries and veins have distinct origins and are formed by different mechanisms. This information may help develop better cell therapies for coronary artery disease.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures

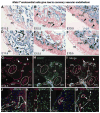
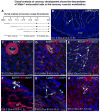
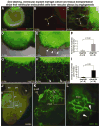

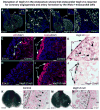

Comment in
-
An endothelial contribution to coronary vessels.Cell. 2012 Nov 21;151(5):932-4. doi: 10.1016/j.cell.2012.11.004. Cell. 2012. PMID: 23178115
References
-
- Carmeliet P, Ferreira V, Breier G, Pollefeyt S, Kieckens L, Gertsenstein M, Fahrig M, Vandenhoeck A, Harpal K, Eberhardt C, et al. Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nature. 1996;380:435–439. - PubMed
-
- Chang CP, Neilson JR, Bayle JH, Gestwicki JE, Kuo A, Stankunas K, Graef IA, Crabtree GR. A field of myocardial-endocardial NFAT signaling underlies heart valve morphogenesis.[see comment] Cell. 2004;118:649–663. - PubMed
-
- Chen JW, Zhou B, Yu QC, Shin SJ, Jiao K, Schneider MD, Baldwin HS, Bergelson JM. Cardiomyocyte-specific deletion of the coxsackievirus and adenovirus receptor results in hyperplasia of the embryonic left ventricle and abnormalities of sinuatrial valves. Circ Res. 2006;98:923–930. - PubMed
-
- de la Pompa JL, Timmerman LA, Takimoto H, Yoshida H, Elia AJ, Samper E, Potter J, Wakeham A, Marengere L, Langille BL, et al. Role of the NF-ATc transcription factor in morphogenesis of cardiac valves and septum.[see comment] Nature. 1998;392:182–186. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

