Tackling the minority: sulfate-reducing bacteria in an archaea-dominated subsurface biofilm
- PMID: 23178669
- PMCID: PMC3578563
- DOI: 10.1038/ismej.2012.133
Tackling the minority: sulfate-reducing bacteria in an archaea-dominated subsurface biofilm
Abstract
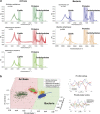
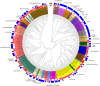
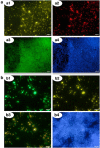
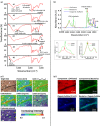
Archaea are usually minor components of a microbial community and dominated by a large and diverse bacterial population. In contrast, the SM1 Euryarchaeon dominates a sulfidic aquifer by forming subsurface biofilms that contain a very minor bacterial fraction (5%). These unique biofilms are delivered in high biomass to the spring outflow that provides an outstanding window to the subsurface. Despite previous attempts to understand its natural role, the metabolic capacities of the SM1 Euryarchaeon remain mysterious to date. In this study, we focused on the minor bacterial fraction in order to obtain insights into the ecological function of the biofilm. We link phylogenetic diversity information with the spatial distribution of chemical and metabolic compounds by combining three different state-of-the-art methods: PhyloChip G3 DNA microarray technology, fluorescence in situ hybridization (FISH) and synchrotron radiation-based Fourier transform infrared (SR-FTIR) spectromicroscopy. The results of PhyloChip and FISH technologies provide evidence for selective enrichment of sulfate-reducing bacteria, which was confirmed by the detection of bacterial dissimilatory sulfite reductase subunit B (dsrB) genes via quantitative PCR and sequence-based analyses. We further established a differentiation of archaeal and bacterial cells by SR-FTIR based on typical lipid and carbohydrate signatures, which demonstrated a co-localization of organic sulfate, carbonated mineral and bacterial signatures in the biofilm. All these results strongly indicate an involvement of the SM1 euryarchaeal biofilm in the global cycles of sulfur and carbon and support the hypothesis that sulfidic springs are important habitats for Earth's energy cycles. Moreover, these investigations of a bacterial minority in an Archaea-dominated environment are a remarkable example of the great power of combining highly sensitive microarrays with label-free infrared imaging.
Figures

References
-
- Adler HH, Kerr PF. Variations in infrared spectra, molecular symmetry and site symmetry of sulfate minerals. Am Mineralogist. 1965;50:132–147.
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

