The chemical interactome space between the human host and the genetically defined gut metabotypes
- PMID: 23178670
- PMCID: PMC3603391
- DOI: 10.1038/ismej.2012.141
The chemical interactome space between the human host and the genetically defined gut metabotypes
Abstract
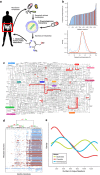
The bacteria that colonize the gastrointestinal tracts of mammals represent a highly selected microbiome that has a profound influence on human physiology by shaping the host's metabolic and immune system activity. Despite the recent advances on the biological principles that underlie microbial symbiosis in the gut of mammals, mechanistic understanding of the contributions of the gut microbiome and how variations in the metabotypes are linked to the host health are obscure. Here, we mapped the entire metabolic potential of the gut microbiome based solely on metagenomics sequencing data derived from fecal samples of 124 Europeans (healthy, obese and with inflammatory bowel disease). Interestingly, three distinct clusters of individuals with high, medium and low metabolic potential were observed. By illustrating these results in the context of bacterial population, we concluded that the abundance of the Prevotella genera is a key factor indicating a low metabolic potential. These metagenome-based metabolic signatures were used to study the interaction networks between bacteria-specific metabolites and human proteins. We found that thirty-three such metabolites interact with disease-relevant protein complexes several of which are highly expressed in cells and tissues involved in the signaling and shaping of the adaptive immune system and associated with squamous cell carcinoma and bladder cancer. From this set of metabolites, eighteen are present in DrugBank providing evidence that we carry a natural pharmacy in our guts. Furthermore, we established connections between the systemic effects of non-antibiotic drugs and the gut microbiome of relevance to drug side effects and health-care solutions.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

