Structural analysis of the oligomeric states of Helicobacter pylori VacA toxin
- PMID: 23178866
- PMCID: PMC3612943
- DOI: 10.1016/j.jmb.2012.11.020
Structural analysis of the oligomeric states of Helicobacter pylori VacA toxin
Abstract
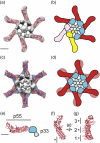
Helicobacter pylori is a Gram-negative bacterium that colonizes the human stomach and contributes to peptic ulceration and gastric adenocarcinoma. H. pylori secretes a pore-forming exotoxin known as vacuolating toxin (VacA). VacA contains two distinct domains, designated p33 and p55, and assembles into large "snowflake"-shaped oligomers. Thus far, no structural data are available for the p33 domain, which is essential for membrane channel formation. Using single-particle electron microscopy and the random conical tilt approach, we have determined the three-dimensional structures of six VacA oligomeric conformations at ~15-Å resolution. The p55 domain, composed primarily of β-helical structures, localizes to the peripheral arms, while the p33 domain consists of two globular densities that localize within the center of the complexes. By fitting the VacA p55 crystal structure into the electron microscopy densities, we have mapped inter-VacA interactions that support oligomerization. In addition, we have examined VacA variants/mutants that differ from wild-type (WT) VacA in toxin activity and/or oligomeric structural features. Oligomers formed by VacA∆6-27, a mutant that fails to form membrane channels, lack an organized p33 central core. Mixed oligomers containing both WT and VacA∆6-27 subunits also lack an organized core. Oligomers formed by a VacA s2m1 chimera (which lacks cell-vacuolating activity) and VacAΔ301-328 (which retains vacuolating activity) each contain p33 central cores similar to those of WT oligomers. By providing the most detailed view of the VacA structure to date, these data offer new insights into the toxin's channel-forming component and the intermolecular interactions that underlie oligomeric assembly.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures




References
-
- Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;1:1311–1315. - PubMed
-
- Suerbaum S, Michetti P. Helicobacter pylori infection. N. Engl. J. Med. 2002;347:1175–1186. - PubMed
-
- Atherton JC. The pathogenesis of Helicobacter pylori-induced gastro-duodenal diseases. Annu. Rev. Pathol. 2006;1:63–96. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

