Database proton NMR chemical shifts for RNA signal assignment and validation
- PMID: 23180050
- PMCID: PMC3555346
- DOI: 10.1007/s10858-012-9683-9
Database proton NMR chemical shifts for RNA signal assignment and validation
Abstract
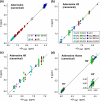
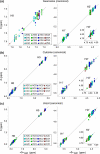
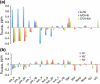
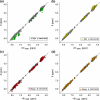
The Biological Magnetic Resonance Data Bank contains NMR chemical shift depositions for 132 RNAs and RNA-containing complexes. We have analyzed the (1)H NMR chemical shifts reported for non-exchangeable protons of residues that reside within A-form helical regions of these RNAs. The analysis focused on the central base pair within a stretch of three adjacent base pairs (BP triplets), and included both Watson-Crick (WC; G:C, A:U) and G:U wobble pairs. Chemical shift values were included for all 4(3) possible WC-BP triplets, as well as 137 additional triplets that contain one or more G:U wobbles. Sequence-dependent chemical shift correlations were identified, including correlations involving terminating base pairs within the triplets and canonical and non-canonical structures adjacent to the BP triplets (i.e. bulges, loops, WC and non-WC BPs), despite the fact that the NMR data were obtained under different conditions of pH, buffer, ionic strength, and temperature. A computer program (RNAShifts) was developed that enables convenient comparison of RNA (1)H NMR assignments with database predictions, which should facilitate future signal assignment/validation efforts and enable rapid identification of non-canonical RNA structures and RNA-ligand/protein interaction sites.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

