Vivaldi: visualization and validation of biomacromolecular NMR structures from the PDB
- PMID: 23180575
- PMCID: PMC3618379
- DOI: 10.1002/prot.24213
Vivaldi: visualization and validation of biomacromolecular NMR structures from the PDB
Abstract
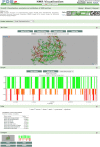
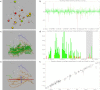
We describe Vivaldi (VIsualization and VALidation DIsplay; http://pdbe.org/vivaldi), a web-based service for the analysis, visualization, and validation of NMR structures in the Protein Data Bank (PDB). Vivaldi provides access to model coordinates and several types of experimental NMR data using interactive visualization tools, augmented with structural annotations and model-validation information. The service presents information about the modeled NMR ensemble, validation of experimental chemical shifts, residual dipolar couplings, distance and dihedral angle constraints, as well as validation scores based on empirical knowledge and databases. Vivaldi was designed for both expert NMR spectroscopists and casual non-expert users who wish to obtain a better grasp of the information content and quality of NMR structures in the public archive.
Copyright © 2013 Wiley Periodicals, Inc.
Figures

References
-
- Berman HM. The Protein Data Bank: a historical perspective. Acta Crystallogr A. 2008;64:88–95. - PubMed
-
- Bernstein FC, Koetzle TF, Williams GJB, Meyer EF, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. Protein Data Bank—computer-based archival file for macromolecular structures. J Mol Biol. 1977;112:535–542. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

