Evolution of animal and plant dicers: early parallel duplications and recurrent adaptation of antiviral RNA binding in plants
- PMID: 23180579
- PMCID: PMC3563972
- DOI: 10.1093/molbev/mss263
Evolution of animal and plant dicers: early parallel duplications and recurrent adaptation of antiviral RNA binding in plants
Abstract
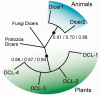
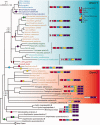
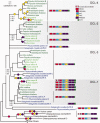
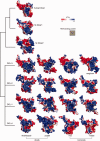
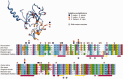
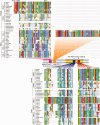
RNA interference (RNAi) is a eukaryotic molecular system that serves two primary functions: 1) gene regulation and 2) protection against selfish elements such as viruses and transposable DNA. Although the biochemistry of RNAi has been detailed in model organisms, very little is known about the broad-scale patterns and forces that have shaped RNAi evolution. Here, we provide a comprehensive evolutionary analysis of the Dicer protein family, which carries out the initial RNA recognition and processing steps in the RNAi pathway. We show that Dicer genes duplicated and diversified independently in early animal and plant evolution, coincident with the origins of multicellularity. We identify a strong signature of long-term protein-coding adaptation that has continually reshaped the RNA-binding pocket of the plant Dicer responsible for antiviral immunity, suggesting an evolutionary arms race with viral factors. We also identify key changes in Dicer domain architecture and sequence leading to specialization in either gene-regulatory or protective functions in animal and plant paralogs. As a whole, these results reveal a dynamic picture in which the evolution of Dicer function has driven elaboration of parallel RNAi functional pathways in animals and plants.
Figures
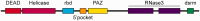
Similar articles
-
Increased Affinity for RNA Targets Evolved Early in Animal and Plant Dicer Lineages through Different Structural Mechanisms.Mol Biol Evol. 2017 Dec 1;34(12):3047-3063. doi: 10.1093/molbev/msx187. Mol Biol Evol. 2017. PMID: 29106606 Free PMC article.
-
Recurrent adaptation in RNA interference genes across the Drosophila phylogeny.Mol Biol Evol. 2011 Feb;28(2):1033-42. doi: 10.1093/molbev/msq284. Epub 2010 Oct 22. Mol Biol Evol. 2011. PMID: 20971974
-
Genetic Insight into the Domain Structure and Functions of Dicer-Type Ribonucleases.Int J Mol Sci. 2021 Jan 9;22(2):616. doi: 10.3390/ijms22020616. Int J Mol Sci. 2021. PMID: 33435485 Free PMC article. Review.
-
The evolution of core proteins involved in microRNA biogenesis.BMC Evol Biol. 2008 Mar 25;8:92. doi: 10.1186/1471-2148-8-92. BMC Evol Biol. 2008. PMID: 18366743 Free PMC article.
-
[Progress of studies on Dicer structure and function].Yi Chuan. 2008 Dec;30(12):1550-6. doi: 10.3724/sp.j.1005.2008.01550. Yi Chuan. 2008. PMID: 19073568 Review. Chinese.
Cited by
-
Identification and Analysis of microRNAs in Chlorella sorokiniana Using High-Throughput Sequencing.Genes (Basel). 2020 Sep 25;11(10):1131. doi: 10.3390/genes11101131. Genes (Basel). 2020. PMID: 32992970 Free PMC article.
-
Plant protein-coding gene families: Their origin and evolution.Front Plant Sci. 2022 Sep 7;13:995746. doi: 10.3389/fpls.2022.995746. eCollection 2022. Front Plant Sci. 2022. PMID: 36160967 Free PMC article. Review.
-
Uncovering a Complex Virome Associated with the Cacao Pathogens Ceratocystis cacaofunesta and Ceratocystis fimbriata.Pathogens. 2023 Feb 9;12(2):287. doi: 10.3390/pathogens12020287. Pathogens. 2023. PMID: 36839559 Free PMC article.
-
The miRNA biogenesis in marine bivalves.PeerJ. 2016 Mar 7;4:e1763. doi: 10.7717/peerj.1763. eCollection 2016. PeerJ. 2016. PMID: 26989613 Free PMC article.
-
Roles of Dicer-Like Proteins 2 and 4 in Intra- and Intercellular Antiviral Silencing.Plant Physiol. 2017 Jun;174(2):1067-1081. doi: 10.1104/pp.17.00475. Epub 2017 Apr 28. Plant Physiol. 2017. PMID: 28455401 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

