Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells
- PMID: 23180760
- PMCID: PMC3531057
- DOI: 10.1093/nar/gks1111
Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells
Abstract
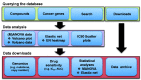
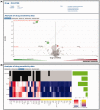
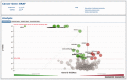
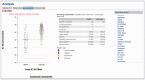
Alterations in cancer genomes strongly influence clinical responses to treatment and in many instances are potent biomarkers for response to drugs. The Genomics of Drug Sensitivity in Cancer (GDSC) database (www.cancerRxgene.org) is the largest public resource for information on drug sensitivity in cancer cells and molecular markers of drug response. Data are freely available without restriction. GDSC currently contains drug sensitivity data for almost 75 000 experiments, describing response to 138 anticancer drugs across almost 700 cancer cell lines. To identify molecular markers of drug response, cell line drug sensitivity data are integrated with large genomic datasets obtained from the Catalogue of Somatic Mutations in Cancer database, including information on somatic mutations in cancer genes, gene amplification and deletion, tissue type and transcriptional data. Analysis of GDSC data is through a web portal focused on identifying molecular biomarkers of drug sensitivity based on queries of specific anticancer drugs or cancer genes. Graphical representations of the data are used throughout with links to related resources and all datasets are fully downloadable. GDSC provides a unique resource incorporating large drug sensitivity and genomic datasets to facilitate the discovery of new therapeutic biomarkers for cancer therapies.
Figures
References
-
- Druker BJ, Guilhot F, O’Brien SG, Gathmann I, Kantarjian H, Gattermann N, Deininger MW, Silver RT, Goldman JM, Stone RM, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N. Engl. J. Med. 2006;355:2408–2417. - PubMed
-
- Stratton MR. Exploring the genomes of cancer cells: progress and promise. Science. 2011;331:1553–1558. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

