Collaborative biocuration--text-mining development task for document prioritization for curation
- PMID: 23180769
- PMCID: PMC3504477
- DOI: 10.1093/database/bas037
Collaborative biocuration--text-mining development task for document prioritization for curation
Abstract
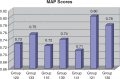
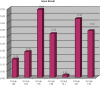
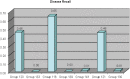
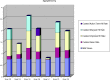
The Critical Assessment of Information Extraction systems in Biology (BioCreAtIvE) challenge evaluation is a community-wide effort for evaluating text mining and information extraction systems for the biological domain. The 'BioCreative Workshop 2012' subcommittee identified three areas, or tracks, that comprised independent, but complementary aspects of data curation in which they sought community input: literature triage (Track I); curation workflow (Track II) and text mining/natural language processing (NLP) systems (Track III). Track I participants were invited to develop tools or systems that would effectively triage and prioritize articles for curation and present results in a prototype web interface. Training and test datasets were derived from the Comparative Toxicogenomics Database (CTD; http://ctdbase.org) and consisted of manuscripts from which chemical-gene-disease data were manually curated. A total of seven groups participated in Track I. For the triage component, the effectiveness of participant systems was measured by aggregate gene, disease and chemical 'named-entity recognition' (NER) across articles; the effectiveness of 'information retrieval' (IR) was also measured based on 'mean average precision' (MAP). Top recall scores for gene, disease and chemical NER were 49, 65 and 82%, respectively; the top MAP score was 80%. Each participating group also developed a prototype web interface; these interfaces were evaluated based on functionality and ease-of-use by CTD's biocuration project manager. In this article, we present a detailed description of the challenge and a summary of the results.
Figures








References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

