Organization of the influenza virus replication machinery
- PMID: 23180774
- PMCID: PMC3578580
- DOI: 10.1126/science.1227270
Organization of the influenza virus replication machinery
Abstract
Influenza virus ribonucleoprotein complexes (RNPs) are central to the viral life cycle and in adaptation to new host species. RNPs are composed of the viral genome, viral polymerase, and many copies of the viral nucleoprotein. In vitro cell expression of all RNP protein components with four of the eight influenza virus gene segments enabled structural determination of native influenza virus RNPs by means of cryogenic electron microscopy (cryo-EM). The cryo-EM structure reveals the architecture and organization of the native RNP, defining the attributes of its largely helical structure and how polymerase interacts with nucleoprotein and the viral genome. Observations of branched-RNP structures in negative-stain electron microscopy and their putative identification as replication intermediates suggest a mechanism for viral replication by a second polymerase on the RNP template.
Figures
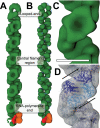



Comment in
-
Biochemistry. Visualizing the influenza genome.Science. 2012 Dec 21;338(6114):1545-6. doi: 10.1126/science.1231588. Epub 2012 Nov 22. Science. 2012. PMID: 23180772 No abstract available.
References
-
- Fields BN, Knipe DM, Howley PM. Fields' Virology. Wolters Kluwer Health/Lippincott Williams & Wilkins; 2007.
-
- Hatta M, Gao P, Halfmann P, Kawaoka Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science. 2001;293:1840–1842. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

