Crystal structure of the calcium release-activated calcium channel Orai
- PMID: 23180775
- PMCID: PMC3695727
- DOI: 10.1126/science.1228757
Crystal structure of the calcium release-activated calcium channel Orai
Abstract
The plasma membrane protein Orai forms the pore of the calcium release-activated calcium (CRAC) channel and generates sustained cytosolic calcium signals when triggered by depletion of calcium from the endoplasmic reticulum. The crystal structure of Orai from Drosophila melanogaster, determined at 3.35 angstrom resolution, reveals that the calcium channel is composed of a hexameric assembly of Orai subunits arranged around a central ion pore. The pore traverses the membrane and extends into the cytosol. A ring of glutamate residues on its extracellular side forms the selectivity filter. A basic region near the intracellular side can bind anions that may stabilize the closed state. The architecture of the channel differs markedly from other ion channels and gives insight into the principles of selective calcium permeation and gating.
Figures

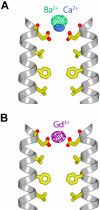
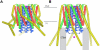
Comment in
-
CRACking the structure of Orai: new work reveals the intramolecular properties of a store-operated, calcium influx channel.Channels (Austin). 2013 Mar-Apr;7(2):71-3. doi: 10.4161/chan.24163. Epub 2013 Mar 1. Channels (Austin). 2013. PMID: 23502496 Free PMC article. No abstract available.
References
-
- Feske S, et al. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature. 2006;441:179. doi:10.1038/nature04702 Medline. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

