The International Nucleotide Sequence Database Collaboration
- PMID: 23180798
- PMCID: PMC3531182
- DOI: 10.1093/nar/gks1084
The International Nucleotide Sequence Database Collaboration
Abstract
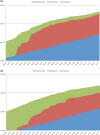
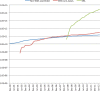
The International Nucleotide Sequence Database Collaboration (INSDC; http://www.insdc.org), one of the longest-standing global alliances of biological data archives, captures, preserves and provides comprehensive public domain nucleotide sequence information. Three partners of the INSDC work in cooperation to establish formats for data and metadata and protocols that facilitate reliable data submission to their databases and support continual data exchange around the world. In this article, the INSDC current status and update for the year of 2012 are presented. Among discussed items of international collaboration meeting in 2012, BioSample database and changes in submission are described as topics.
Figures