Decoding human cytomegalovirus
- PMID: 23180859
- PMCID: PMC3817102
- DOI: 10.1126/science.1227919
Decoding human cytomegalovirus
Abstract
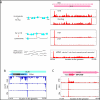
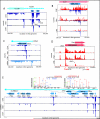
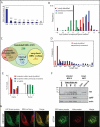
The human cytomegalovirus (HCMV) genome was sequenced 20 years ago. However, like those of other complex viruses, our understanding of its protein coding potential is far from complete. We used ribosome profiling and transcript analysis to experimentally define the HCMV translation products and follow their temporal expression. We identified hundreds of previously unidentified open reading frames and confirmed a fraction by means of mass spectrometry. We found that regulated use of alternative transcript start sites plays a broad role in enabling tight temporal control of HCMV protein expression and allowing multiple distinct polypeptides to be generated from a single genomic locus. Our results reveal an unanticipated complexity to the HCMV coding capacity and illustrate the role of regulated changes in transcript start sites in generating this complexity.
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

