Genome-wide association study of survival in patients with pancreatic adenocarcinoma
- PMID: 23180869
- PMCID: PMC3816124
- DOI: 10.1136/gutjnl-2012-303477
Genome-wide association study of survival in patients with pancreatic adenocarcinoma
Abstract
Background and objective: Survival of patients with pancreatic adenocarcinoma is limited and few prognostic factors are known. We conducted a two-stage genome-wide association study (GWAS) to identify germline variants associated with survival in patients with pancreatic adenocarcinoma.
Methods: We analysed overall survival in relation to single nucleotide polymorphisms (SNPs) among 1005 patients from two large GWAS datasets, PanScan I and ChinaPC. Cox proportional hazards regression was used in an additive genetic model with adjustment for age, sex, clinical stage and the top four principal components of population stratification. The first stage included 642 cases of European ancestry (PanScan), from which the top SNPs (p≤10(-5)) were advanced to a joint analysis with 363 additional patients from China (ChinaPC).
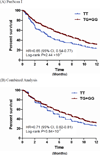
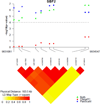
Results: In the first stage of cases of European descent, the top-ranked loci were at chromosomes 11p15.4, 18p11.21 and 1p36.13, tagged by rs12362504 (p=1.63×10(-7)), rs981621 (p=1.65×10(-7)) and rs16861827 (p=3.75×10(-7)), respectively. 131 SNPs with p≤10(-5) were advanced to a joint analysis with cases from the ChinaPC study. In the joint analysis, the top-ranked SNP was rs10500715 (minor allele frequency, 0.37; p=1.72×10(-7)) on chromosome 11p15.4, which is intronic to the SET binding factor 2 (SBF2) gene. The HR (95% CI) for death was 0.74 (0.66 to 0.84) in PanScan I, 0.79 (0.65 to 0.97) in ChinaPC and 0.76 (0.68 to 0.84) in the joint analysis.
Conclusions: Germline genetic variation in the SBF2 locus was associated with overall survival in patients with pancreatic adenocarcinoma of European and Asian ancestry. This association should be investigated in additional large patient cohorts.
Keywords: Cancer Genetics; Molecular Epidemiology; Pancreatic Cancer.
Conflict of interest statement
Figures

Similar articles
-
Germline polymorphisms and survival of lung adenocarcinoma patients: a genome-wide study in two European patient series.Int J Cancer. 2015 Mar 1;136(5):E262-71. doi: 10.1002/ijc.29195. Epub 2014 Sep 19. Int J Cancer. 2015. PMID: 25196286
-
Relevance of Sp Binding Site Polymorphism in WWOX for Treatment Outcome in Pancreatic Cancer.J Natl Cancer Inst. 2016 Feb 8;108(5):djv387. doi: 10.1093/jnci/djv387. Print 2016 May. J Natl Cancer Inst. 2016. PMID: 26857392 Free PMC article.
-
A genome-wide association study of overall survival in pancreatic cancer patients treated with gemcitabine in CALGB 80303.Clin Cancer Res. 2012 Jan 15;18(2):577-84. doi: 10.1158/1078-0432.CCR-11-1387. Epub 2011 Dec 5. Clin Cancer Res. 2012. PMID: 22142827 Free PMC article.
-
Lack of replication of seven pancreatic cancer susceptibility loci identified in two Asian populations.Cancer Epidemiol Biomarkers Prev. 2013 Feb;22(2):320-3. doi: 10.1158/1055-9965.EPI-12-1182. Epub 2012 Dec 18. Cancer Epidemiol Biomarkers Prev. 2013. PMID: 23250936
-
Epidemiology and Inherited Predisposition for Sporadic Pancreatic Adenocarcinoma.Hematol Oncol Clin North Am. 2015 Aug;29(4):619-40. doi: 10.1016/j.hoc.2015.04.009. Hematol Oncol Clin North Am. 2015. PMID: 26226901 Free PMC article. Review.
Cited by
-
Germline Features Associated with Immune Infiltration in Solid Tumors.Cell Rep. 2020 Mar 3;30(9):2900-2908.e4. doi: 10.1016/j.celrep.2020.02.039. Cell Rep. 2020. PMID: 32130895 Free PMC article.
-
Implementation of individualised polygenic risk score analysis: a test case of a family of four.BMC Med Genomics. 2022 Oct 3;15(Suppl 3):207. doi: 10.1186/s12920-022-01331-8. BMC Med Genomics. 2022. PMID: 36192731 Free PMC article.
-
Association of gastrointestinal gland cancer susceptibility loci with esophageal carcinoma among the Chinese Han population: a case-control study.Tumour Biol. 2016 Feb;37(2):1627-33. doi: 10.1007/s13277-015-3945-6. Epub 2015 Aug 26. Tumour Biol. 2016. PMID: 26304507
-
SurvivalGWAS_SV: software for the analysis of genome-wide association studies of imputed genotypes with "time-to-event" outcomes.BMC Bioinformatics. 2017 May 19;18(1):265. doi: 10.1186/s12859-017-1683-z. BMC Bioinformatics. 2017. PMID: 28525968 Free PMC article.
-
Identification and Validation of a Biomarker Signature in Patients With Resectable Pancreatic Cancer via Genome-Wide Screening for Functional Genetic Variants.JAMA Surg. 2019 Jun 1;154(6):e190484. doi: 10.1001/jamasurg.2019.0484. Epub 2019 Jun 19. JAMA Surg. 2019. PMID: 30942874 Free PMC article.
References
-
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. - PubMed
-
- Ferlay J, Shin HR, Bray F, et al. GLOBOCAN 2008 v1.2, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 10 [Internet] Lyon, France: International Agency for Research on Cancer; 2010. [accessed on 30/03/2012]. Available from: http://globocan.iarc.fr.
-
- Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362:1605–1617. - PubMed
-
- Wu C, Xu B, Yuan P, et al. Genome-wide interrogation identifies YAP1 variants associated with survival of small-cell lung cancer patients. Cancer Res. 2010;70:9721–9729. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA73790/CA/NCI NIH HHS/United States
- P30 CA016087/CA/NCI NIH HHS/United States
- R37 CA70867/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- P30 ES000260/ES/NIEHS NIH HHS/United States
- R01 CA047988/CA/NCI NIH HHS/United States
- R01 CA105069/CA/NCI NIH HHS/United States
- N01 RC037004/RC/CCR NIH HHS/United States
- U01 AG018033/AG/NIA NIH HHS/United States
- R01 CA082729/CA/NCI NIH HHS/United States
- P01 CA087969/CA/NCI NIH HHS/United States
- CA 97193/CA/NCI NIH HHS/United States
- HHSN261201000006C/CP/NCI NIH HHS/United States
- P30 CA015704/CA/NCI NIH HHS/United States
- HL 26490/HL/NHLBI NIH HHS/United States
- P01 CA055075/CA/NCI NIH HHS/United States
- R37 CA070867/CA/NCI NIH HHS/United States
- 5U01AG018033/AG/NIA NIH HHS/United States
- R01 HL034595/HL/NHLBI NIH HHS/United States
- N01 WH022110/WH/WHI NIH HHS/United States
- G1000143/MRC_/Medical Research Council/United Kingdom
- UM1 CA173640/CA/NCI NIH HHS/United States
- HL 34595/HL/NHLBI NIH HHS/United States
- R01 CA124908/CA/NCI NIH HHS/United States
- CA 40360/CA/NCI NIH HHS/United States
- 14136/CRUK_/Cancer Research UK/United Kingdom
- CA 34944/CA/NCI NIH HHS/United States
- G0401527/MRC_/Medical Research Council/United Kingdom
- R01CA098661/CA/NCI NIH HHS/United States
- P01 CA87969/CA/NCI NIH HHS/United States
- R01CA034588/CA/NCI NIH HHS/United States
- R01 CA098661/CA/NCI NIH HHS/United States
- P30 CA015083/CA/NCI NIH HHS/United States
- NCI K07 CA140790/CA/NCI NIH HHS/United States
- P01 CA55075/CA/NCI NIH HHS/United States
- R01 HL026490/HL/NHLBI NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- R01 CA82729/CA/NCI NIH HHS/United States
- K07 CA140790/CA/NCI NIH HHS/United States
- R01 CA040360/CA/NCI NIH HHS/United States
- CA105069/CA/NCI NIH HHS/United States
- N01 CN045165/CN/NCI NIH HHS/United States
- HHSN261200800001C/RC/CCR NIH HHS/United States
- UM1 CA182910/CA/NCI NIH HHS/United States
- P50 CA127003/CA/NCI NIH HHS/United States
- L30 CA130644/CA/NCI NIH HHS/United States
- R01 CA097193/CA/NCI NIH HHS/United States
- R01 CA034944/CA/NCI NIH HHS/United States
- HHSN261200800001E/CA/NCI NIH HHS/United States
- ES000260/ES/NIEHS NIH HHS/United States
- P30CA016087/CA/NCI NIH HHS/United States
