Pathway choice in DNA double strand break repair: observations of a balancing act
- PMID: 23181949
- PMCID: PMC3557175
- DOI: 10.1186/2041-9414-3-9
Pathway choice in DNA double strand break repair: observations of a balancing act
Abstract
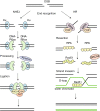
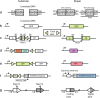
Proper repair of DNA double strand breaks (DSBs) is vital for the preservation of genomic integrity. There are two main pathways that repair DSBs, Homologous recombination (HR) and Non-homologous end-joining (NHEJ). HR is restricted to the S and G2 phases of the cell cycle due to the requirement for the sister chromatid as a template, while NHEJ is active throughout the cell cycle and does not rely on a template. The balance between both pathways is essential for genome stability and numerous assays have been developed to measure the efficiency of the two pathways. Several proteins are known to affect the balance between HR and NHEJ and the complexity of the break also plays a role. In this review we describe several repair assays to determine the efficiencies of both pathways. We discuss how disturbance of the balance between HR and NHEJ can lead to disease, but also how it can be exploited for cancer treatment.
Figures
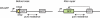

References
-
- Friedberg ECW, Siede GC, Wood RD, Schultz RA, Ellenberger T. DNA repair and mutagenesis. Washington, USA: ASM press; 2006.
-
- Buck D, Malivert L, de Chasseval R, Barraud A, Fondaneche MC, Sanal O, Plebani A, Stephan JL, Hufnagel M, le Deist F. et al.Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
