A cyclin without cyclin-dependent kinases: cyclin F controls genome stability through ubiquitin-mediated proteolysis
- PMID: 23182110
- PMCID: PMC3597434
- DOI: 10.1016/j.tcb.2012.10.011
A cyclin without cyclin-dependent kinases: cyclin F controls genome stability through ubiquitin-mediated proteolysis
Abstract
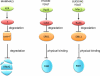
Cell cycle transitions are driven by the periodic oscillations of cyclins, which bind and activate cyclin-dependent kinases (CDKs) to phosphorylate target substrates. Cyclin F uses a substrate recruitment strategy similar to that of the other cyclins, but its associated catalytic activity is substantially different. Indeed, cyclin F is the founding member of the F-box family of proteins, which are the substrate recognition subunits of Skp1-Cul1-F-box protein (SCF) ubiquitin ligase complexes. Here, we discuss cyclin F function and recently identified substrates of SCF(cyclin)(F) involved in deoxyribonucleotide triphosphate (dNTP) production, centrosome duplication, and spindle formation. We highlight the relevance of cyclin F in controlling genome stability through ubiquitin-mediated proteolysis and the implications for cancer development.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures

References
-
- Satyanarayana A, Kaldis P. Mammalian cell-cycle regulation: several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene. 2009;28(33):2925–39. - PubMed
-
- Kraus B. A novel cyclin gene (CCNF) in the region of the polycystic kidney disease gene (PKD1) Genomics. 1994;24(1):27–33. - PubMed
-
- Bai C. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86(2):263–74. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

